|
To view this email as a web page, click here. |
 |
|
Welcome
We hope to see many of you at the World HUPO meeting in Florida, so please stop by and visit us at Booth #307.
Check that your search parameters are fully optimized, especially if you have new sources of data.
This month's highlighted publication shows a method for non-invasive extraction and identification of proteins from works of art.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month deals with the ever-expanding NCBI nr database.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
Do you need to optimize your search parameters?
If some aspect of your data has changed, perhaps due to new instrumentation, your search results may be improved by re-optimizing your search parameters.
Here is a simple workflow for determining the best parameters for your particular data type:
- Review the experimental background to determine basics such as sample treatment, species, instrumentation, fragmentation method, labeling, and enrichment.
- Perform a basic search with wider than normal mass tolerances and minimal modifications using an 'average' subset of the data.
- Determine the mass tolerance by looking at the precursor peptide mass errors for the top protein hits. Also review the mass error graph for high mass peptides with high scores and estimate a suitable tolerance for the MS/MS ions.
- Find common modifications by re-searching the data as an error tolerant search with these tolerances. The goal is to determine if there are unusual numbers of any particular modification or non-specific cleavages.
- Run the optimized search and include a decoy in the search. Once the search completes, check the False Discovery Rate and adjust to 1%.
Go here to read more details about this optimization process.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Minimally Invasive and Portable Method for the Identification of Proteins in Ancient Paintings
Paola Cicatiello, Georgia Ntasi, Manuela Rossi, Gennaro Marino, Paola Giardina, and Leila Birolo
Anal. Chem., 2018, 90, 10128-10133
The authors have developed a method for capturing and identifying proteins from works of art in order to understand aging and deterioration in historical objects. Invasiveness of analytical measurements is a central issue in conservation and restoration of cultural heritage goods, and this method demonstrated no significant surface changes at the microscopic level.
The method uses a cellulose acetate sheet that is coated with Vmh2 hydrophobin, which then non-covalently immobilizes the trypsin enzyme. Dampened sheets are then gently put in contact with the artwork, removing the proteins and generating the peptides. Extraction of the peptides from the sheet is followed by MALDI-TOF MS and MS/MS analysis to identify the proteins.
Eight different historical samples were investigated, one from the detached frescoes by Buonamico Buffalmacco (XIII century) from the Monumental Cemetery in Pisa, and seven unknown painting from the XIX to XX centuries. The results on the Buffalmacco frescoes were in agreement with previously published results.
|
 |
 |
 |
 |
|
Mascot Tip
Do you have the NCBI nr comprehensive protein database (NCBIProt) on your Mascot Server? If so, please note that NCBI have recently added a couple of entries that are longer than the default limit of 50,000 residues. If you want to include these entries in searches, increase this limit before you next update the nr Fasta:
- Configuration editor
- Configuration options
- Change MaxSequenceLen to 80000
- Apply
Don't set MaxSequenceLen a lot higher than it needs to be because this wastes RAM.
Whether you really want to search these entries is worth thinking about. They are PWL93229 and PWL95011, both from an unpublished paper on the human gut metagenome and annotated as "Derived by automated computational analysis using gene prediction method: GeneMarkS+." Possibly a literal example of GIGO! You might prefer to leave the setting at 50,000 residues so that these two entries are skipped.
NCBI nr is now ridiculously large. The downloadable file is 38GB and unpacks to 94GB. Bringing it online is slow and takes up a great deal of disk space. In most cases, there are better choices of database, such as a subset of GenBank for the organism of interest or a Uniprot complete proteome.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
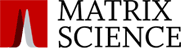 |
 |
|
|