|
To view this email as a web page, click here. |
 |
|
Happy New Year!
We have some suggestions for distinguishing between different 1 Da modifications.
This month's highlighted publication gives an overview of identifying unexpected or novel modifications.
If you have a recent publication that you would like us to consider for an upcoming Newsletter, please
send us a PDF or a URL.
Mascot tip of the month describes how Report Builder in the Protein Family Summary can be used to create a customised table of protein hits.
Please have a read and feel free to contact us if you have any comments or questions. |
|
|
|
 |
 |
 |
|
One Dalton, many possibilities
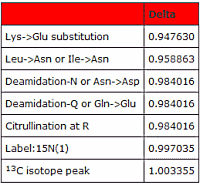
When you encounter a peptide whose mass has increased by 1 Dalton, it is often challenging to determine which of the many possible modifications has occurred.
A common initial thought is that the peak picking software simply selected the 13C peak. With high mass accuracy this can be sorted out since, at mass 1500, the difference between the 13C peptide mass and deamidation is 13ppm. Additionally, the MS/MS match should show some fragments at +1 for the deamidation case, but none for the 13C peptide.
Things get more difficult when the modified residue is near the amino terminus and the predominant fragments are y ions, so that there are few or no +1 peaks to be observed (or, vice versa.)
Deamidation versus citrullination is particularly thorny since the mass shift is identical. The decision may depend on a combination of site localisation, the neutral loss of isocyanic acid, and the immonium ion of citrulline, as well as comparison with reference spectra.
And then there is the confusing case of Label:15N(1) often being present in error tolerant search results. Go here to read more about unravelling this as well as other one Dalton dilemmas.
|
 |
 |
 |
 |
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here please send us a PDF or a URL.
|
|
|
Identification of Unexpected Protein Modifications by Mass Spectrometry-Based Proteomics
Shiva Ahmadi and Dominic Winter
In: Wang X., Kuruc M. (eds) Functional Proteomics. Methods in Molecular Biology, vol 1871. Humana Press, New York, NY
The authors provide a strategy for the discovery of unexpected modifications in complex samples by bottom-up LC-MS/ MS. While expected modifications are readily identified by selecting them as variable modifications, the identification of unknown modifications is less straightforward and requires further data analysis and validation.
They describe in great detail an experimental workflow which includes:
- Parallel searches for the identification of known modifications at unexpected amino acids
- Error tolerant searches for modifications unexpected in the sample but known to the community
- Mass tolerant searches to discover novel modifications.
Additionally, they suggest follow-up verification of identified modifications in the initial dataset and targeted experiments using synthetic peptides.
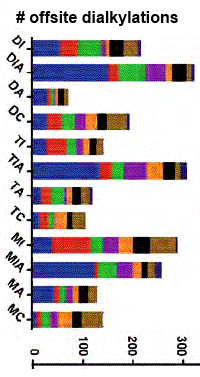
Procedures were illustrated by analysing a cytosolic fraction of HeLa cells. With LC/MS/MS, the authors identified and quantified a large number of side reactions by monomers and dimers of the alkylation reagents (see figure at right). They also determined that alkylation of peptides with iodine-containing reagents results in a loss of the side chain of methionine. Some 22 notes provide helpful tips on sample preparation, data acquisition, and data processing.
|
 |
 |
 |
 |
|
Mascot Tip
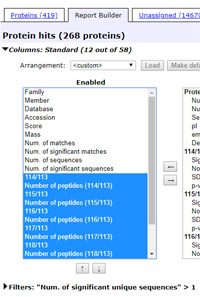
Report Builder in the Protein Family Summary report is a powerful way to create a customised table of protein hits. You can choose which columns to include and their order, filter out proteins that are of no interest, such as one-hit wonders, and export the table in CSV format directly to Excel.
To see how Report Builder works, load a typical report for a search of iTRAQ 8plex data by clicking on this link. Expand the Sensitivity and FDR section of the header and set the FDR to 1% for peptide sequences (not PSMs). Once the report has re-loaded, switch to the Report Builder tab.
Filter out one-hit wonders by expanding the Filters section and selecting Num. of significant unique sequences > 1 then choosing Filter. You should now have 268 high-confidence proteins listed.
To select and re-order columns, expand the Columns section. Use the Arrangement drop down list to load a saved arrangement or, for this exercise, choose <custom> to configure a new table design.
Under Enabled, select emPAI and Description and move them to Available. Under Available, for each ratio, select the ratio value and the number of peptides and move all 14 items to Enabled. (To select multiple items at once, control+click on individual items or shift+click a range.)
Most likely, the new block of items is now at the top of the Enabled list. To move them to the end of the list, which will be the right hand side of the table, with all items selected, use the down arrow below the list. Finally, choose Apply.
You can sort the table on any column by clicking on the header. If this was your in-house Mascot Server, and you thought this column arrangement might be useful in future, you could save it by expanding the Column section once more, entering a suitable name, and choosing Save. Saved column arrangements are initially private to your Mascot security login, but can made global as explained here.
|
 |
 |
 |
 |
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Please contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|
 |
 |
|
|