|
To view this email as a web page, click here. |
|
|
|
Welcome
We are developing novel precursor detection for spectrum-centric peptide identification from DIA spectra.
This month's highlighted publication reconstructs mediaeval monastic diets from teeth and bone.
Microsoft has announced the end of Windows 10 mainstream support on 14 October 2025, which affects Mascot Server 2.6 and 2.7.
|
|
|
|
|
|
 |
|
Mascot: The trusted reference standard for protein identification by mass spectrometry for 25 years
|
Get a quote
|
 |
|
|
|
Precursor detection in Mascot DIA
|
|
|
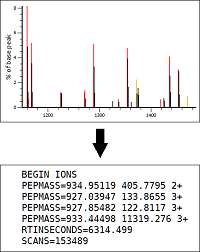
We're working on a universal spectrum-centric solution for the analysis of DIA data, called Mascot DIA.
It includes a novel algorithm for precursor detection, which will be implemented in the upcoming Mascot Distiller 3.0.
In data-independent acquisition, the instrument fragments all precursors within a selected isolation window.
One of the software challenges is to figure out the masses of those precursors, as the raw file only stores the isolation window boundaries.
The conventional approach is to make a list of all theoretical precursors from a sequence database or spectral library, and try all masses that fit the isolation window.
Instead, we are developing a novel algorithm that infers precursor masses directly from the observed peaks in the MS/MS scan.
Briefly, the algorithm selects pairs of complementary ions from the MS/MS scan and adds their masses.
This yields a potential precursor MH+ mass plus an additional proton, which is converted to precursor m/z.
If a sufficient number of peak pairs are found for a precursor m/z within the isolation window, it's added as a candidate.
Once all peak pairs have been processed, the top candidates are selected for database searching.
This approach has several advantages over conventional methods:
- You don't need a library of precursor masses and associated retention times.
- Supports any enzyme and variable modifications.
- Precursor mass and charge can be determined even from centroided data.
- Largely independent of fragmentation efficiency.
- Works with any complementary ion series, such as b/y and c/z.
More details and a worked example are available in our blog.
|
|
|
|
 |
|
|
|
Featured publication using Mascot
Here we highlight a recent interesting and important publication that employs Mascot for protein identification, quantitation, or characterization. If you would like one of your papers highlighted here, please send us a PDF or a URL.
|
|
|
Reconstructing medieval diets through the integration of stable isotope and proteomic analyses from two European burial sites
A. Pedergana, J. Grossmann, R. Turck, A. Goujon, F. Rühli, S. Wilkin, P. Eppenberger, C. Lehn
Scientific Reports, 15, Article number: 26442 (2025), doi:10.1038/s41598-025-10103-0
Stable isotope data from human teeth can provide information on an individual's diet in childhood, while ribs can be used to determine the average dietary composition in adulthood.
However, it only provides a broad overview.
Dental calculus is the mineralised biofilm on tooth surfaces, which may store a variety of lipids and proteins from plant and animal sources.
The authors analysed dental calculus, dentin and bone samples collected from two medieval monastic cemeteries in Germany (Dalheim) and Switzerland.
The goal was to test whether dietary inferences obtained from dental calculus are related to stable isotope data from collagen.
Stable isotope data reveal that the Dalheim population relied primarily on proteins from terrestrial animals, alongside the consumption of mainly C3 plants.
While isotope analyses did not provide clear evidence of fish or legume consumption, proteomic data confirmed the presence of both in the diet.
Identified taxa included common wheat, barley, millet, green peas and European perch.
However, the recovery rate of dietary proteins from calculus samples from the two sites was fairly low.
A total of 16 dietary proteins (34 unique peptides) were recovered from 15 individuals.
Although the two analysis methods are complementary, the authors note the importance of considering protein structure in the differential preservation of dietary proteins in dental calculus.
|
|
|
|
 |
|
|
|
Windows 10 support is ending
|
|
|
Microsoft has announced the end of Windows 10 mainstream support on 14 October 2025.
If you are running Mascot Server 2.8 or 3.x on Windows 10, we recommend updating to Windows 11.
However, if you have Mascot Server 2.6 or 2.7, note that these versions are not compatible with Windows 11 due to a fault in a third-party Perl module.
One option is to continue running Windows 10, although you will no longer get security updates for the operating system.
If you are required to update Windows, then you will also need to update to Mascot Server 3.1, which is fully compatible with Windows 11.
To find out which version you have, go to your local Mascot home page and select Database Status.
The version number is shown at the top of the page.
This is an excellent time to update the software.
Since 1 July, Mascot Server is available in three new sizes (Small, Medium, Large), and the cost to update to the latest version has been greatly reduced.
A version update also includes 1 year of Premium Support with remote support and guided installation/migration.
Please email us at support@matrixscience.com for more details and a quote.
|
 |
|
|
 |
|
|
|
About Matrix Science
Matrix Science is a provider of bioinformatics tools to proteomics researchers and scientists, enabling the rapid, confident identification and quantitation of proteins. Mascot continues to be cited by over 2000 publications every year. Our software products fully support data from mass spectrometry instruments made by Agilent, Bruker, Sciex, Shimadzu, Thermo Scientific, and Waters.
Get a quote
|
|
|
You can also contact us or one of our marketing partners for more information on how you can power your proteomics with Mascot.
|





|
|
|
|
|