NCBI nr in Mascot Server 2.8.1
Mascot Server 2.8.01 patch was released in March 2022. One of the big improvements is optimising the compression speed of the NCBI nr database, available as the NCBIprot predefined definition. We’ve greatly decreased the time it takes to bring the database online, as well as removed an inadvertent limitation on database size. The patch is available to download on the technical support page.
NCBI nr size
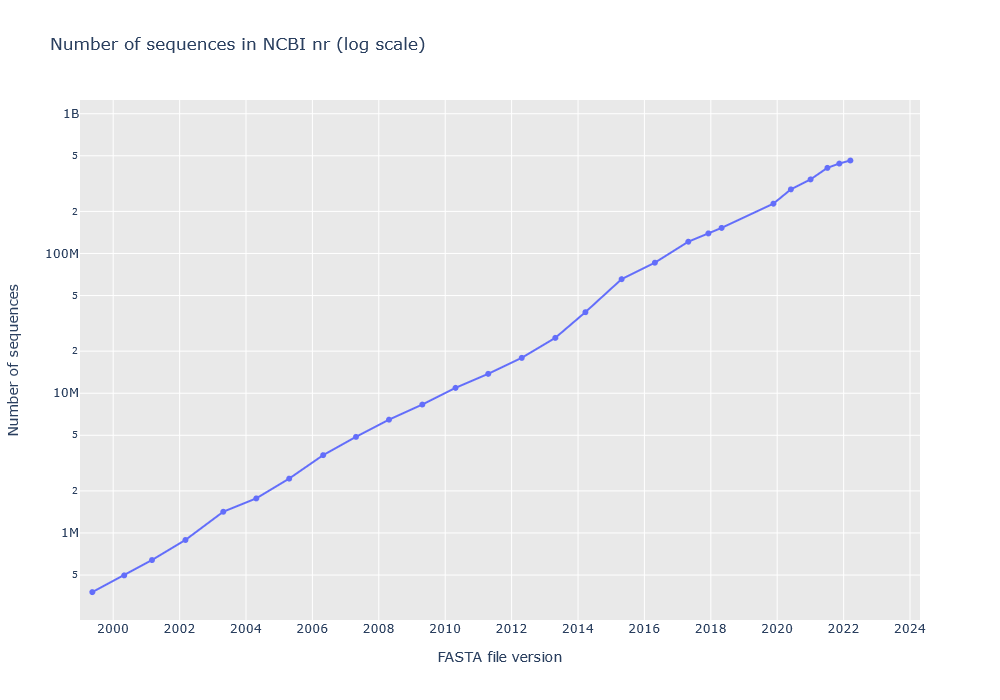
NCBI nr doubles in size roughly every two years. The database includes protein sequences from several other databases, including RefSeq and GenBank. The main driver for new sequences seems to be RefSeq.
There is no official graph for the number of protein sequences in nr. The time series below has been created from search logs on our public website. The FASTA file version and sequence count is saved in the header of the Mascot results file, and the public website has had NCBInr or NCBIprot as a searchable database since 1999.
The y axis is log scale and the slope has stayed the same for 20 years. It should reach 1 billion sequences by 2024 or 2025. As you can imagine, a database this large can hit all kinds of hardware and software limits.
Database size limit – solved
When Mascot brings a FASTA database online, it creates several compressed index files whose purpose is to allow fast streaming through the protein sequences. One of the index files, .t00, stores the taxonomy IDs for each database entry. NCBI nr is one of the few databases where a protein sequence may have multiple accessions, so each entry may have multiple taxonomy IDs. Mascot uses a data structure that stores the file position (aka offset) of each protein in the index files. When it advances to the next database entry, it fast forwards the file handles based on the offset values.
Unfortunately, the data structure used a 32-bit integer as the .t00 file offset. When the index file is over 4GB in size, the file position wraps around and causes non-deterministic behaviour. The most common symptom is, the database search never finishes and may eventually crash.
Based on test results with NCBIprot, Mascot 2.8.0 and earlier can handle 339 million sequences but not 410 million sequences. We didn’t spend much time finding exactly where the limit is between the two numbers; determining the root cause and fixing the fault had higher priority! To be precise, the bug is triggered by the number of accessions, not sequences. nr currently has approximately 1.8 accessions per database entry, so earlier Mascot versions work with FASTA files with up to about 650 million accessions. A database without taxonomy parse rule isn’t subject to the limit, as it won’t use a .t00 index.
Mascot 2.8.01 now uses a 64-bit file offset for the .t00 file, so the maximum .t00 file size is 8 exabytes (8192 petabytes). Assuming NCBI nr continues doubling every two years, it may reach 1 trillion sequences by 2044. The .t00 file would be 16TB, well below the new 64-bit limit. The FASTA file for 1 trillion sequences could be around 470TB, so it’s quite likely NCBI will split the file by kingdom or phylum before then and we’ll never reach the new .t00 size limit.
Compression speed – solved
The other issue is the time taken to create the index files and bring NCBIprot online. Many years ago when nr was smaller, compression time was a linear function of database size. In recent years, the relationship has broken down.
The nr FASTA file is supplemented by prot.accession2taxid, which maps protein accessions to taxonomy IDs. Mascot’s NCBIprot definition uses a smaller table, prot.av2taxid, which we create from prot.accession2taxid. During database compression, most of the index files are created sequentially and need little RAM. However, nr taxonomy lookups are different. The prot.av2taxid table must be available for random access lookup, because the primary and secondary accessions in the FASTA file are not in the same order as prot.accession2taxid or prot.av2taxid.
Mascot 2.8.0 and earlier create a read-only constant database, .cdb, for prot.av2taxid, which provides the fast random access lookup. The table is divided into several sequential .cdb files, which are then memory mapped. The division strategy was good enough for many years, until prot.av2taxid reached a certain size and the constant database was split in far too many parts. In computer science terms, the random access lookup was meant to be O(1) (constant time) but slowly became O(n) (linear time) over the years, causing a severe disk bottleneck.
Mascot 2.8.01 uses a different strategy and now horizontally partitions prot.av2taxid by the first character of the accession string. The two-level lookup should be O(1) again and the improvement is borne out in benchmarking results. The .cdb files continue to be memory mapped. The more available RAM you have, the more of the lookup files the operating system can keep cached in memory, and the faster the lookups become.
Hardware recommendation
The previous NCBI nr tips article briefly discussed disk performance but didn’t give any hard numbers. We benchmarked several hardware configurations, which are summarised in the table below.
| Type | CPU | RAM | Disk | Platform | NCBIprot compression 2.8.0 and earlier, 339M sequences |
NCBIprot compression 2.8.01, 440M sequences |
|---|---|---|---|---|---|---|
| Server | Intel Xeon 3.4GHz | 128GB | RAID10, 10k rpm HDDs | Linux | 5h | 6h |
| Workstation | Intel Core i7 3.2GHz | 64GB | SSD and HDD | Windows | 240h | 21h |
| Virtual machine | Intel Xeon 2.5GHz | 32GB | Virtualised disk on RAID50 host, 10k rpm HDDs | Linux | cancelled, predicted to take >1 week | 23-25h |
| Laptop | Intel Core i7 2.4GHz | 16GB | SSD and HDD | Windows | cancelled after 1 week, infeasible | 52h |
The 2.8.01 benchmarks used nr FASTA from November 2021, which contained 440 million sequences. The 2.8.0 benchmarks used a January 2021 FASTA, which had 339 million sequences, as the later file cannot be brought online in the previous version. The new version succeeds in every case, while Mascot 2.8.0 was very slow on the workstation and unusable on the virtual machine and the laptop.
The two most important factors are amount of RAM and disk speed. As you can see, a RAID10 array with 10k rpm disks gives the shortest possible compression time, and there isn’t much difference between the old and new versions. Decreasing the amount of available RAM decreases compression speed, until compression becomes disk bound and slow. There is little difference between HDD and SSD compared to RAID. We recommend having enough RAM for twice the size of the prot.av2taxid file. Currently 32GB is plenty, but you should plan for 64GB. On non-virtualised systems and if nothing else is using much RAM, it is possible to bring NCBIprot online in a couple days with less RAM, like the laptop example in the table.
Using NCBIprot in Mascot 2.7 and earlier
If you want to use NCBIprot in Mascot 2.7 and earlier, there are two options.
First option is to download the latest FASTA file and split it in at least two parts. Set up each part as a separate database. Instructions are in sequence database setup help. The advantage is having the latest version available. The downside is, you will need serious hardware. You should have at least 128GB RAM and a RAID5 or RAID10 array.
Alternatively, you can set up an earlier version of the FASTA file. Anything before July 2021 should work with Mascot 2.7 and earlier. The advantage is being able to use the predefined configuration and a single FASTA file. The downside is, you are stuck with the old version. If you’d like to use this option and don’t have a suitable FASTA file, please contact us.
Keywords: database manager, Fasta, pc hardware