SCIEX instruments
SCIEX instruments for proteomics are currently divided into five families:
- QTOF systems (e.g. ZenoTOF 7600)
- QTRAP systems (e.g. QTRAP 6500+)
- Triple Quad systems (e.g. Triple Quad 7500)
- TripleTOF systems (e.g. TripleTOF 6500+)
- X-Series TOF (e.g. X500B QTOF)
The above instruments all use SCIEX Analyst® acquisition software, producing files with a .wiff or .wiff2 extension.
There are a number of legacy instruments. Data in the T2D format produced by older SCIEX MALDI instruments can be processed with Mascot Distiller and is documented under SCIEX Data Explorer.
Mascot Distiller
Mascot Distiller can be used to browse and process wiff files into high quality peak lists that can be saved or submitted direct to a Mascot Server for searching. The search results can be imported back into Distiller for further examination or used as the basis for quantitation. If the optional Mascot Daemon Toolbox is installed, these processes can be automated using Mascot Daemon.
Mascot Distiller does not currently support files in wiff2 format. SCIEX OS version 2.1 and later produce files in wiff format compatible with Mascot Distiller.
For ZenoTOF instruments, Mascot Server 2.8 and later ship with a suitable EAD instrument definition.
Data-dependent acquisition (DDA)
Mascot Distiller makes full use of DDA spectra from all SCIEX instruments, as long as the results are presented in wiff format (not wiff2). Distiller has full support for identification and quantitation from DDA data.
SWATH and DIA
Mascot Distiller can open SWATH runs (Sequential Window Acquisition of all Theoretical Mass Spectra) saved in with format, but it does not currently support deconvoluting the spectra.
If the spectra are acquired using narrow, fixed DIA windows (<8 m/z), then current Distiller versions produce reasonable peak lists. Full support is planned in a future Mascot Distiller release.
Mascot Daemon
Mascot Daemon can be used to process batches of raw files by choosing Mascot Distiller as the data import filter.
Mascot Distiller requires the optional Mascot Daemon Toolbox to allow the Distiller libraries to be called from Mascot Daemon. When this toolbox is active, Mascot Distiller will appear automatically on the list of data import filters in Daemon.
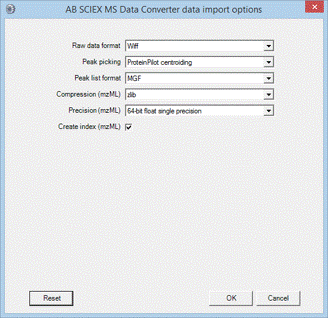
SCIEX MS Data Converter
SCIEX MS Data Converter is a free download from SCIEX. It supports both wiff files and runs on Windows 10 and later. Output can be a simple MGF peak list or an mzML file containing a fairly complete representation of the raw data.
It may be useful to perform ProteinPilot peak picking using SCIEX MS Data Converter and open it in Mascot Distiller (for example, quantitation or de novo identification). To do this, convert the wiff file to an mzML file. The command line will be similar to this:
AB_SCIEX_MS_Converter.exe WIFF C:\tmp\fraction_42.wiff -proteinpilot MZML C:\tmp\fraction_42.mzml
Remember to use double quotes around the file paths if they include any spaces. When the mzML file is opened in Distiller, you need to choose processing options that uncentroid the survey scans but take the existing centroid values from the MS/MS scans.
The converter also allows profile data to be exported to an mzML file. You can open the exported mzML file in Distiller, but it is usually better to work with the original Wiff file.
SCIEX MS Data Converter can be used as a data import filter in Mascot Daemon. This enables batch automation of peak picking and search submission.
MSConvert
MSConvert is a component of ProteoWizard that converts between various file formats. It has a large number of options and can be configured as a data import filter in Mascot Daemon. Note that the input file format is not specified; msconvert auto-detects the format.
Some versions of msconvert, when converting wiff to MGF, are unable to output precursor charge in the MGF file. For these versions, it is important to select a default peptide charge when submitting the search. If the peptide charge control is hidden (Mascot Server 3.0 and later), the default is taken from the SearchSubmitDefaultPeptideCharge option.
All trade marks and service marks belonging to SCIEX are hereby acknowledged.