Exercise DIS2: answers
- Try to find a peptide match using denovo for the summed spectrum of scans 1616-1624
De novo using the suggested settings will give solutions similar to these. The top two have the same score of 75: [Ai]AG[YG]iCiTiSPK and [Ai]AG[SS|CA]DFiTiSPK.
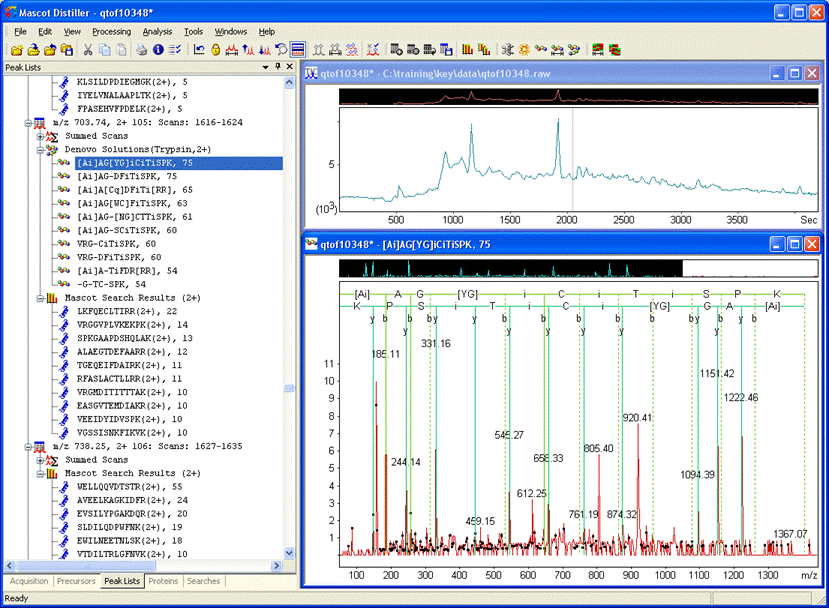
- How can you decide whether to accept any particular match?
The de novo solutions contain quite a lot of ambiguity. Also, why should we hypothesise a novel sequence when it might be a known sequence with a modification. Sometimes, we can find a fit to a database sequence using Blast or MSBlast. The quality of these solutions isn’t good enough for Blast. Right click a de novo solution and try an MSBlast search.
There are no convincing matches from the first solution, [Ai]AG[YG]iCiTiSPK, but the second one gives a number of high scoring matches to human transaldolases
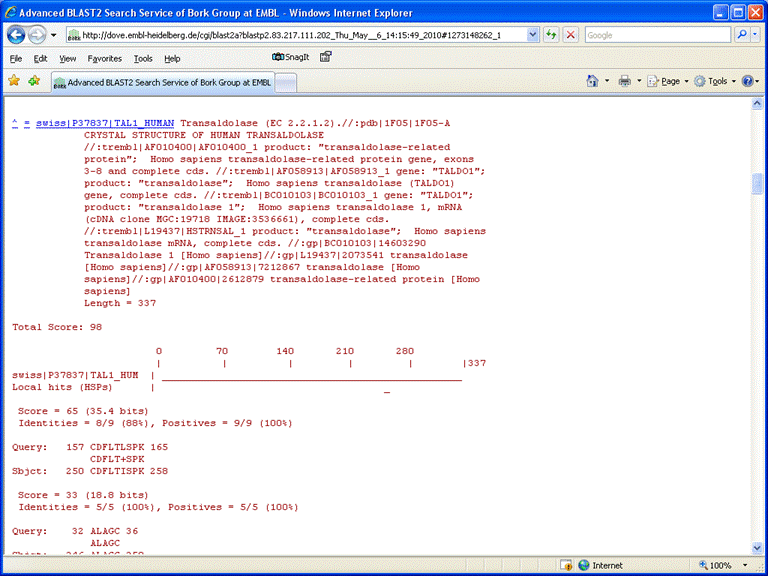
This is one of the strong protein hits found in the original Mascot search. Open the Mascot Protein View report for transaldolase and locate the proposed peptide sequence in the protein. The match is quite convincing:
- de novo: [Ai]AG[SS|CA]DFiTiSPK
- TALDO_HUMAN: K.ALAGCDFLTISPK.L
The only difference is that [SS|CA] in the de novo solution has mapped to C in the database sequence. How could this be explained? You may be able to guess. One possibility is that the cysteine has been modified by acrylamide (71), because the mass of propionamide is the same as Ala.