Exercise SQ3: answers
- Can you get a convincing match and can you explain the mass difference between the database
sequence and the analyte?
De novo using the suggested settings will give solutions similar to those in the screen shot. The top two have the same score of 75: [Ai]AG[YG]iCiTiSPK and [Ai]AG[SS|CA]DFiTiSPK.
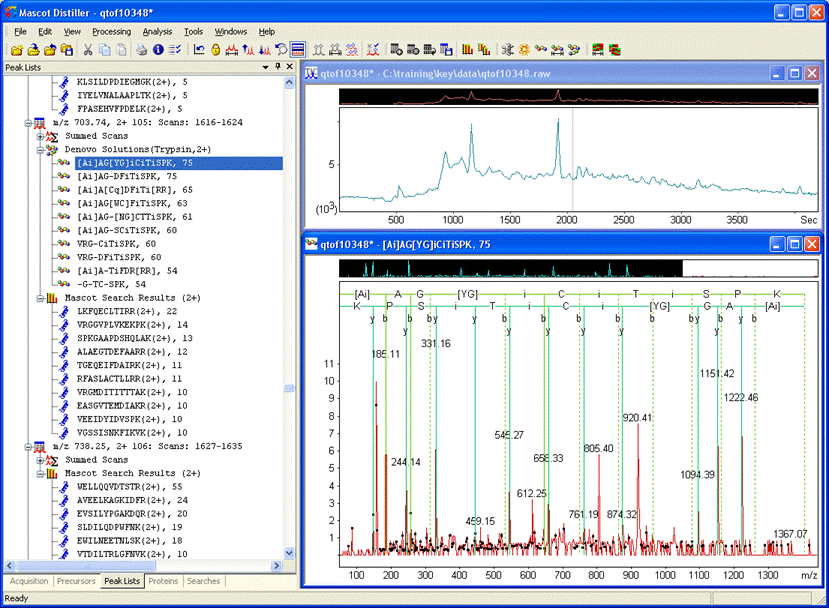
The de novo solutions contain quite a lot of ambiguity. Also, why should we hypothesise a novel sequence when it might be a known sequence with a modification? Right click a de novo solution and try an error tolerant sequence tag search.
Performing an etag search of the second de novo solution in the screen shot above gives two very high scoring matches to transaldolases
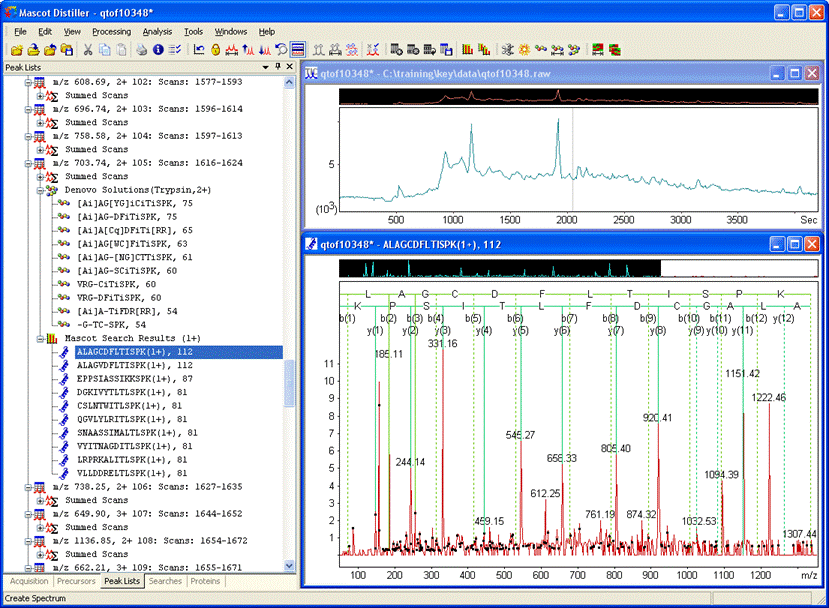
One of these solutions is found in TALDO_HUMAN, which is one of the strong protein hits found in the original Mascot search. Open the Mascot Peptide View report for this match. The mass difference is 70.8 Da on the N-term side of C5. The most likely explanation is that the cysteine has been modified by acrylamide (71).
MS/MS search results
Etag search resultsIf you have time, you could experiment with the Fragment Peptide function in Distiller to see whether there is evidence in the spectrum for the precise site of modification.