Mascot Distiller 20th anniversary
This year is the 25th anniversary of Matrix Science and 25 years of Mascot Server, but it’s also the 20th birthday of Mascot Distiller! Mascot Distiller 1.0 was released in June 2003 and it’s still under active development today. Distiller started as a GUI application for browsing and peak picking native (raw, binary) mass spectrometry data files. Let’s have a look at its story.
The ASMS 2003 presentation that launched Distiller has many vintage screenshots as well as also some historical context. The rationale for Distiller was to provide a single user interface for processing raw data. Many labs – then and now – have instruments from multiple vendors, or at least instruments with different data systems. User interfaces, mouse actions, terminology and icons vary. It’s a real burden to learn and remember the idiosyncracies.
Distiller ships with data providers for all the major vendor formats. Once you learn this single tool, you have a unified interface to process data from any instrument. Equally importantly, the peak picking algorithm chosen for Distiller works with spectra from any instrument. It fits each peak to a calculated isotope distribution, which accurately detects fragment peaks, ignores noise and automatically provides de-isotoping and decharging.
Twenty years later, there are more vendor-independent options for peak picking. ProteoWizard msconvert is the well known solution, and it’s often bundled with free software tools. However, there are only a few solutions that combine vendor-independent peak picking, raw data browsing, database search visualisation and de novo result visualisation in a single package. Mascot Distiller is one of them.
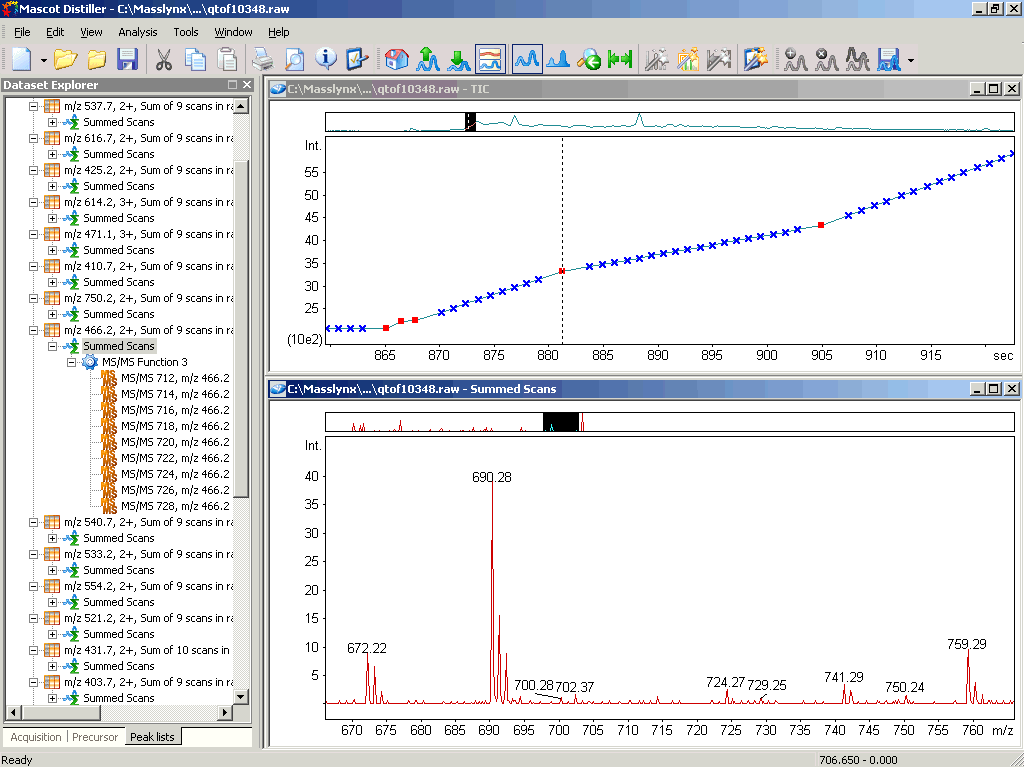
Below is a screenshot of Distiller 2.0 from November 2005. The main GUI elements have not changed much, which is on purpose. We try to avoid changing things without a good reason, as every major change in a user interface forces people to relearn the application.
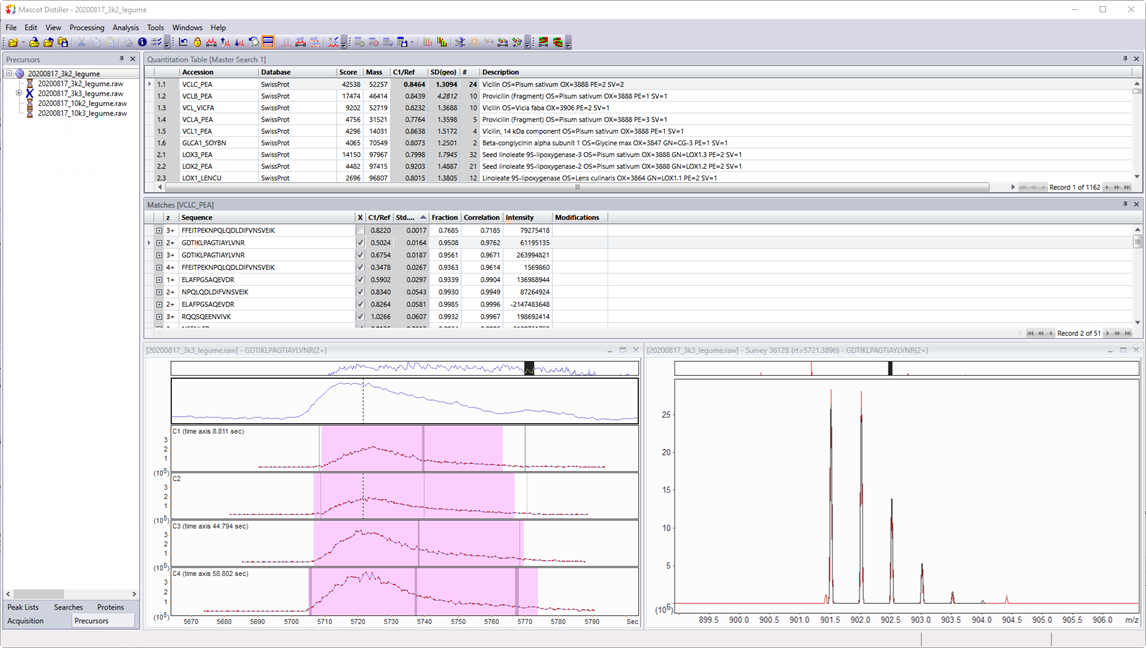
Here’s a screenshot of Distiller 2.8 from two decades later, with an LFQ project, showing Mascot search results and the extracted ion chromatograms (XICs) for the four components.
Mascot Distiller isn’t just for peak picking. The base application was quickly extended with optional toolboxes.
Daemon Toolbox: Launched in Distiller 1.0. Use Mascot Daemon to automate Distiller processing. Initially this was only for automating peak picking, where Daemon uses Distiller as a data import filter. With the introduction of the search toolbox and the quantitation toolbox, Daemon could automate the whole processing from raw data to quantified proteins.
Developer Toolbox: Launched in Distiller 1.0. Allows you to call the Distiller peak picking module from your own Windows applications. The Developer Toolbox provides a uniform Application Programmer Interface to all of the different raw file formats, greatly reducing development time.
Search Toolbox: Launched in Distiller 2.0 in 2005. Provides de novo sequencing and database search integration with Mascot Server. The integration goes both ways: You can peak pick and submit data from Distiller directly to Mascot Server, and when the database search finishes, Distiller imports the results back into the GUI and overlays peptide matches on the MS/MS spectra. This visualisation allows you to quickly evaluate match quality as well as identify any issues with unexplained fragment peaks, mass tolerances, noise levels and so on.
Quantitation Toolbox: Launched in Distiller 2.2 in 2008. Initially supported isobaric quantitation methods like 18O, SILAC and metabolic labeling. Label-free quantitation was added in Distiller 2.3 in 2009. From the beginning, Distiller has emphasised visualisation. Navigate the total ion chromatogram, drill down to MS and MS/MS spectra, examine the XICs, see the effect of changing quantitation filters and so on. This emphasis was unusual fifteen years ago, and it’s still not common among proteomics software packages.
The functionality, convenience, speed and scaling of Distiller quantitation has been vastly improved over the years. The first few versions of Distiller were 32-bit Windows applications. These had a severe RAM limit, maximum 3GB or 4GB depending on Windows version, which made it very difficult to process multi-file projects. Distiller components were incrementally ported to 64-bit, culminating in full 64-bit support in Distiller 2.4. Nowadays, it’s not unusual for workstations to have 64GB or 128GB of RAM, which allows you to create a label-free project with dozens of fractions and replicates. Distiller 2.8 also has vastly improved reporting functionality compared to any earlier version.
We’ve added a new release history page, which includes links to all the related content for more details. The Distiller technical support pages have also been restructured to make it easier to see what information pertains to which version.
Keywords: anniversary, history, Mascot Distiller