What’s new in Mascot Server 3.2 (pre-release draft)
Note: This is a pre-release draft and the details may change before the final release.
Mascot Server is a powerful search engine which uses mass spectrometry data to identify proteins from DNA, RNA and protein sequence databases.
Mascot DIA
Mascot Server and Mascot Distiller can now be used for identifying and quantifying proteins from Data Independent Acquisition (DIA) runs. This is enabled by two key features:
- New in Mascot Distiller 3.0: Novel peak detection for Thermo DIA and SCIEX SWATH (see What’s new in Mascot Distiller 3.0)
- New in Mascot Server 3.2: Noise-resistant scoring for DIA spectra (see below)
Mascot DIA is a spectrum-centric solution and uses the same analysis workflow for DDA and DIA identification and quantitation. Read more in Overview of Mascot DIA.
New: Noise-resistant scoring for Data Independent Acquisition (DIA)
DIA spectra are highly chimeric, containing fragments from multiple precursors as well as unfragmented, overlapping precursor signal. Mascot Server 3.2 introduces a new probabilistic scoring model that is much more noise resistant. Mascot is now able to better ignore peaks from unrelated precursors and dig deeper into the peak list when looking for fragment matches. The peak selection window is also reduced from 100Da (DDA) to 50Da (DIA), allowing Mascot to use twice as many peaks for scoring.
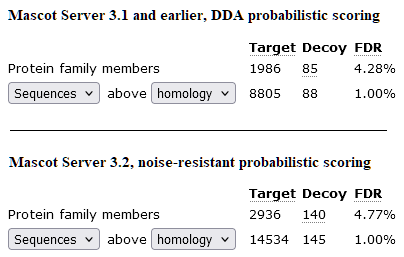
The screenshot below compares sensitivity at 1% sequence FDR for a SCIEX TTOF6600 SWATH run with 99 variable isolation windows (PRIDE project PDX028375). Precursor detection and peak picking was performed by Mascot Distiller 3.0. The DDA probabilistic scoring struggles to identify just 8,806 peptide sequences from the 2h elution. With noise-resistant scoring, Mascot Server 3.2 identifies 14,534 peptide sequences.
Noise-resistant scoring is triggered by the new search parameter DATA_INDEPENDENT_ACQUISITION=1. Mascot Distiller 3.0 sets the search parameter automatically for DIA projects. Peptide scoring for DDA data is unchanged in this version of Mascot Server.
New: Refine merged results with machine learning [DDA, DIA]
Replicate quantitation using Mascot Distiller creates a merged results report, where peptide matches from different runs are combined into a single report. Mascot Server 3.2 can now refine the merged results using machine learning. The refined results can be exported in CSV and mzIdentML formats.
Refining merged results also enables machine learning when creating a quantitation summary report using the Replicate protocol in Mascot Daemon 3.2.
Mascot Server uses a train-then-merge strategy. Individual runs are refined separately, then the refined results are merged. This strategy is required when a DeepLC model is selected, because the predicted retention times are calibrated per run. The train-then-merge strategy is also used when DeepLC is not active.
Improved: Mascot Daemon 3.2
Mascot Daemon is the automation client shipped with Mascot Server. Mascot Server 3.2 ships with the new Mascot Daemon 3.2.
Improved: Core feature set for refining results yields 5-15% more matches [DDA]
We have reviewed the core set of features calculated from PSMs, which are used in refining with machine learning (Percolator). The core feature set was last reviewed in Mascot Server 2.8. On average, the new feature set yields 5-15% more peptide matches across a wide range of DDA runs compared to Mascot Server 3.1.
The previous feature set included four features that are very slow to compute: rawScore, isoSysDM, isoSysDMppm and isoSysDMz. With the new feature set, these dimensions no longer provide useful discrimination between targets and decoys, so they have been removed. This makes refining much faster, often taking half as much time as Mascot Server 3.1.
Improved: Percolator has been updated to version 3.7
Mascot Server 3.1 shipped with Percolator 3.5. In some data sets, enabling an MS2PIP or DeepLC model caused Percolator 3.5 to fail to estimate a PEP (posterior error probability), which led to all peptide matches getting a score of zero and becoming non-significant. The Percolator executable bundled with Mascot Server has been updated to Percolator 3.7, which solves the issue.
Improved: Exporting refined results from client.pl?result_file_mime was unnecessarily slow
The API command client.pl?result_file_mime is used by Thermo Proteome Discoverer as well as Mascot Distiller 2.x. When refining with machine learning is enabled, the command now runs much faster due to removing redundant isValid() checks, up to 6 times faster than Mascot Server 3.1. The impact can be seen by comparing the time to import Mascot results into the client application.
Improved: systemd unit file for Linux
Mascot Server 3.2 includes a new systemd unit file, which integrates the Mascot service (ms-monitor.exe) with the system job control. This replaces the old SysV-init script shipped with previous versions of Mascot.
Other improvements
The release also fixes other minor issues. In total, over 40 change requests and bugs are addressed in this release. The full list is included in the release notes.