Mascot for Data Independent Acquisition (DIA)
Mascot DIA is a spectrum-centric approach for identifying and quantifying proteins from Data Independent Acquisition (DIA) runs. It is built on three products: Mascot Distiller (preprocessing and quantitation), Mascot Server (high-throughput identification) and Mascot Daemon (automation).
The same workflow is used for DDA and DIA processing. Mascot DIA includes novel precursor detection and noise-resistant, probability-based scoring.
Why should I use Mascot for DIA?
Mascot DIA is ideal for untargeted and discovery proteomics, because it has no limits.
- Use any sequence database, from a single proteome to the entire UniProt database (hundreds of millions of sequences).
- Use any variable modifications from Unimod (~1500 mods) or add your own.
- Use any enzyme or search for semi-tryptic or endogenous proteins.
- Use any quantitation method based on MS1 precursor mass shift – label-free quantitation, SILAC, 18O, metabolic.
- High-throughput client-server architecture – any number of client programs can feed data to the same server.
Mascot is ideal for DIA because of its reliability.
- Universal noise-resistant probabilistic scoring works with any instrument and acquisition strategy.
- Universal peak detection works with any instrument and is independent of machine learning.
- Peptide identifications are inherently reliable — Mascot will not report a match unless it has unambiguous fragmentation evidence.
- Protein identifications are inherently reliable — every protein hit must have unique peptide evidence.
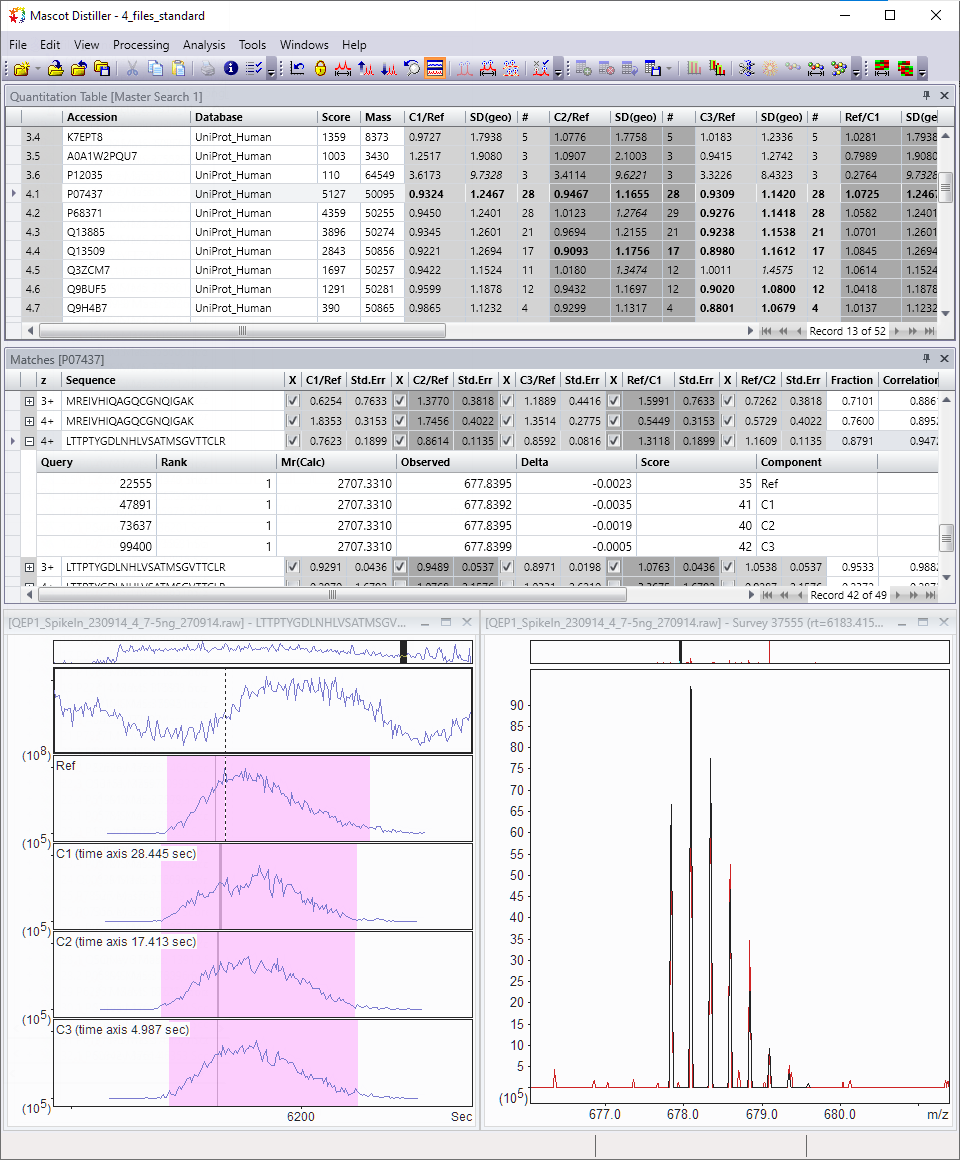
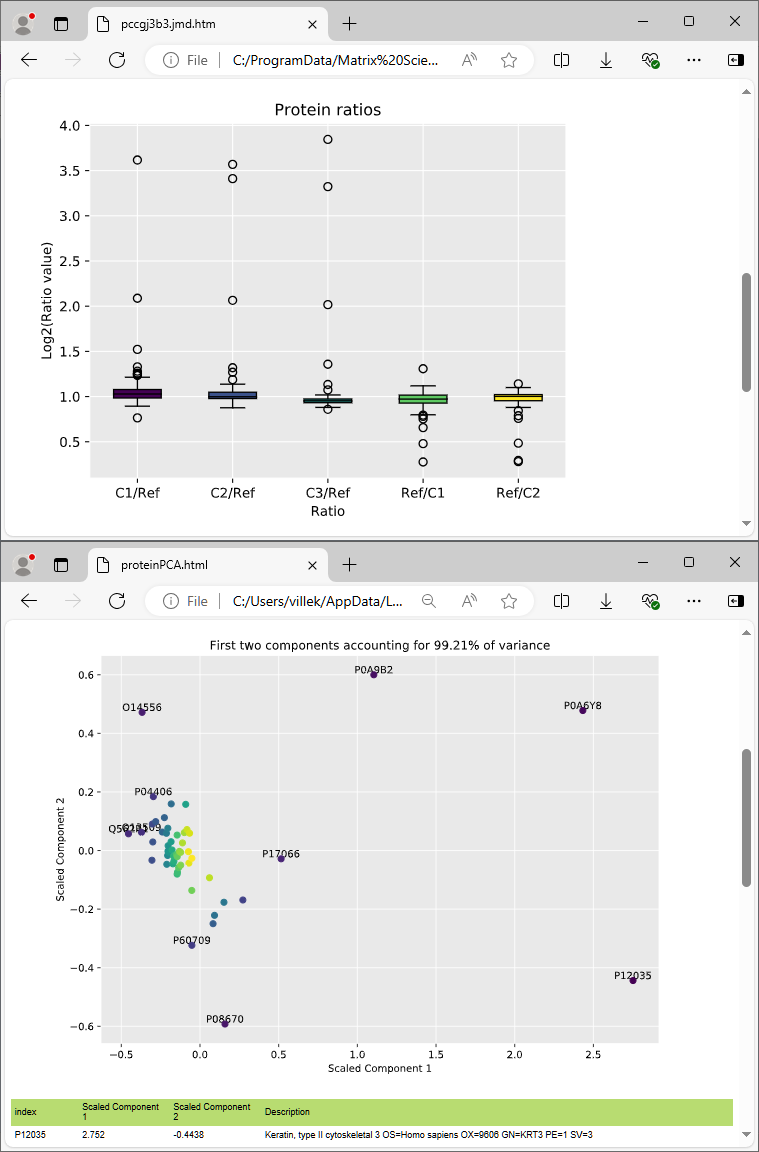
Don’t just take our word for it. Drill down in the interactive reports to inspect the MS/MS spectral evidence, XICs and time alignment results. A number of quantitation and quality reports are included out of the box.
What do I need to get started?
You need Mascot Distiller 3.0. If you have an existing Mascot Distiller 2.x licence, this is a paid update; please contact us for a trial licence.
You also need Mascot Server 3.2 or later. If you have an existing Mascot Server 3.x licence, the update is free provided you have an active support contract. If you have an earlier version or your support has lapsed, please contact us for pricing.
If you need a new licence or wish to evaluate Mascot Server, please contact us or get a quote.
Which instruments are supported?
Mascot DIA currently supports Thermo Orbitrap instruments and SCIEX instruments.
Any Thermo Orbitrap instrument that saves the acquisition in a Thermo Xcalibur raw file is supported. This includes Thermo Orbitrap Astral.
Any SCIEX instrument that saves the acquisition in a wiff file is supported. This includes QTOF systems (e.g. ZenoTOF 7600, 8600) and TripleTOF systems (e.g. 6500+).
Support for Bruker diaPASEF is planned.
The acquisition strategy (SWATH, IDA, etc.) and gradient length do not matter, although you should avoid very short (<15min) gradients. We recommend an isolation window width between 2m/z and 8/mz. Mascot DIA can identify peptides from MS/MS scans with a wider isolation window (>8m/z), but the larger the window, the lower the sensitivity. With wider isolation windows, ion mobility separation is strongly recommended. Isolation windows may be staggered or overlapped in any configuration.
For DDA analysis, Mascot supports all Agilent, Bruker, SCIEX, Shimadzu, Thermo and Waters instruments commonly used in proteomics.
What is the Mascot DIA LFQ workflow?
The Mascot DIA LFQ workflow is exactly the same as the Mascot DDA LFQ workflow:
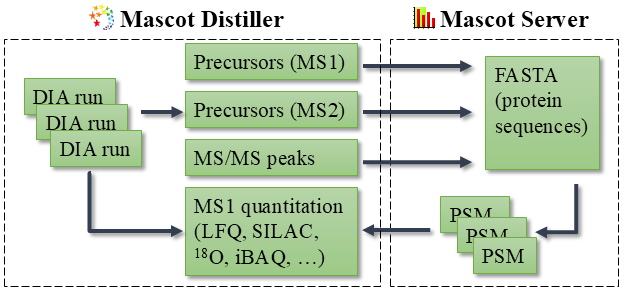
Mascot Distiller reads vendor raw files, processes the MS/MS spectra into peak lists and determines potential precursor masses for each MS/MS spectrum. Precursors are detected from MS1 survey scans as well as from MS2 scans using a novel algorithm. Distiller submits the peak lists to Mascot Server for identification by database searching.
Mascot Server tries to match peptides from a protein sequence database against the observed MS/MS spectrum. Candidate sequences within precursor tolerance are matched independently to the spectrum, allowing multiple precursors to match simultaneously. Mascot calculates a probabilistic match score for each match. At the end of the search, matches are rescored in multiple dimensions using Percolator, and optionally predicted fragment intensities (MS2PIP) and retention time (DeepLC).
Mascot Distiller imports the identified peptides and proteins, and calculates the extracted ion chromatogram (XIC) for each peptide. Multiple runs are aligned in the time dimension based on shared peptide matches. Peptides not identified in the database search may be identified using precursor mass and accurate retention time (“match between runs”). Finally, protein quantitation ratios are calculated from the XICs.
Mascot Daemon automates all steps of preprocessing, database searching and quantitation.
What is spectrum-centric analysis?
DIA software packages can be divided into spectrum-centric and peptide-centric approaches. Mascot DIA is spectrum-centric. The strategies are complementary, with different advantages and disadvantages.
The goal of spectrum-centric identification is to explain as many peaks in the MS/MS spectrum as possible. A peptide is identified if a statistically significant number of theoretical fragment masses match to the observed peaks.
Advantages of spectrum-centric searching:
- Peptide identification requires a strong run of fragment ions, which increases identification reliability and confidence.
- There are no limits on sequence database size, number of variable modifications or enzyme.
- Works with any instrument and any ion series (a, b, c, x, y, z, z+1).
- Uses all the information in the spectrum, including neutral losses, for PTM localisation.
- Mascot DIA does not require a spectral library: peptide identification is independent of machine learning.
Disadvantages:
- Requires a narrow isolation window, ideally 2m/z to 8m/z. With wider windows, or very short gradients, ion mobility separation is highly recommended. The more complex the spectrum, the fewer peptides can be reliably identified.
- If a precursor has no MS1 signal and its mass cannot be inferred from the MS/MS spectrum, then it cannot be identified.
- If a precursor produces only a few fragments, it cannot be confidently identified.
What is peptide-centric analysis?
Mascot DIA is spectrum-centric, but many competing products are peptide-centric.
The goal of peptide-centric identification is to find some evidence for a theoretical precursor in the LC-MS/MS run(s). A peptide is identified by tracing the elution of key fragment peaks.
Advantages of peptide-centric searching:
- Supports wide isolation windows (>8m/z) and potentially very complex MS/MS spectra.
- Theoretical precursors may be detected from sequential MS/MS scans even if there is no MS1 signal.
Disadvantages:
- Peptide-centric tools rely heavily on accurate retention time prediction, because the list of theoretical precursors is built from the expected retention time.
- Peptide-centric tools rely heavily on accurate spectral libraries, whether it’s DDA spectra or predicted spectra from a machine learning model.
- Unless the error rate is carefully controlled, peptide-centric tools may claim to identify a peptide from as few as 3-5 fragments.
- Limited to a handful of PTMs and enzymes.
What kind of PC hardware does Mascot DIA require?
DIA data is inherently complex and data sets are large. DIA database searches are typically 3-5 times larger than DDA database searches.
We recommend an AMD Ryzen 9 or Threadripper processor with 16-32 cores, or equivalent Intel Xeon Gold or Core i9 processor. Processors with more than 32 cores should be avoided, because the clock speed per core is limited due to thermal constraints. You should have 64GB RAM and at least 2TB of fast disk space (NVMe).
Mascot Server and Mascot Distiller run on the CPU and do not require a GPU.
For more details, see PC specification for Mascot Server.