What’s new in Mascot Distiller 3.0 (pre-release draft)
Note: This is a pre-release draft and the details may change before the final release.
Mascot Distiller is a vendor independent application for quantifying proteins and peptides from a Mascot database search.
Product versioning
Mascot Distiller 3.0 is a major release. From now on, a major release is indicated by the first component of the version number. The patch releases to Mascot Distiller 3.0 will have numbers like 3.1 and 3.2; then the next major version after this one will be 4.0.
Mascot DIA
Mascot Server and Mascot Distiller can now be used for identifying and quantifying proteins from Data Independent Acquisition (DIA) runs. This is enabled by two key features:
- New in Mascot Distiller 3.0: Novel peak detection for Thermo DIA and SCIEX SWATH (see below)
- New in Mascot Server 3.2: Noise-resistant scoring for DIA spectra (see What’s new in Mascot Server 3.2)
Mascot DIA is a spectrum-centric solution and uses the same analysis workflow for DDA and DIA identification and quantitation. Read more in Overview of Mascot DIA.
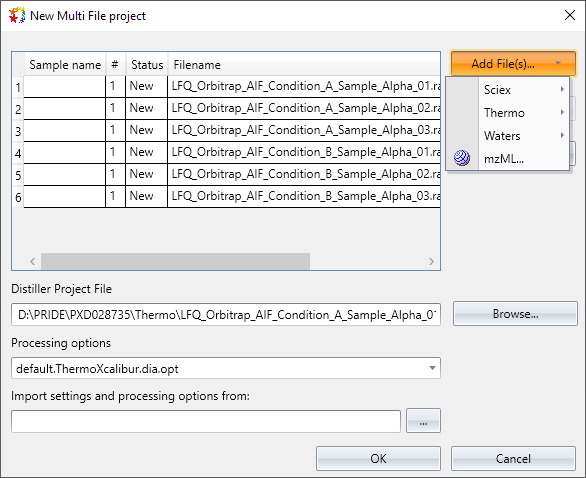
New: Create DIA projects (Thermo and SCIEX instruments)
Mascot Distiller is now able to open and process DIA runs from Thermo and SCIEX instruments, in addition to Waters MSE. To start, create a new DIA single or multi-file project from the File menu:
Select the raw files, and the rest of the steps are the same as with DDA analysis. Any acquisition strategy can be used (SWATH, IDA, fixed/variable isolation windows). Mascot Distiller automatically detects the isolation window specifications and the scan structure from the raw file.
Mascot Distiller 3.0 supports any Thermo Orbitrap instrument that saves raw data in Thermo Xcalibur format, any SCIEX instrument in wiff format and Waters MSE. Support for Bruker diaPASEF is being planned.
We recommend running the samples with narrow isolation windows (<8m/z), or if you use a variable windowing scheme, use narrow isolation windows in the busiest mass regions. Mascot DIA can identify peptides from MS/MS scans with a wider isolation window (>8m/z), but the larger the window, the lower the sensitivity. With wider isolation windows, ion mobility separation is strongly recommended.
New: Thermo Orbitrap Astral support [DDA, DIA]
The Thermo Xcalibur instrument provider has been updated. Mascot Distiller can now process DDA and DIA data acquired on Thermo Orbitrap Astral instruments. Data access of Thermo raw files stored on a network drive has also been greatly improved.
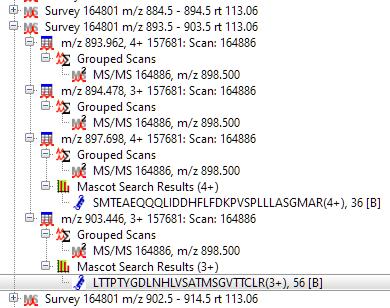
New: Detect precursor masses from MS/MS scans [DIA]
Distiller implements a novel algorithm for chimeric precursor detection for DIA: Precursors are detected from MS1 (survey) scans using the isotopic distribution, as well as deduced from MS/MS scans using complementary ion masses. The algorithm is illustrated in You’ve got to pick a precursor or two.
Precursor detection is universal and works with any instrument. Because it’s data driven and only uses the observed peaks, it’s independent of spectral libraries and machine learning.
New: Identify peptides from DIA data
Mascot Distiller is now able to identify peptides from DIA runs using sequence database searching. Distiller uses the power of Mascot Server to identify peptides. No deconvolution is needed, because Mascot Server identifies peptides directly from the chimeric spectrum. The minimum version for DIA is Mascot Server 3.2.
The peak list tree has been adjusted to group and display chimeric matches in the existing tree structure.
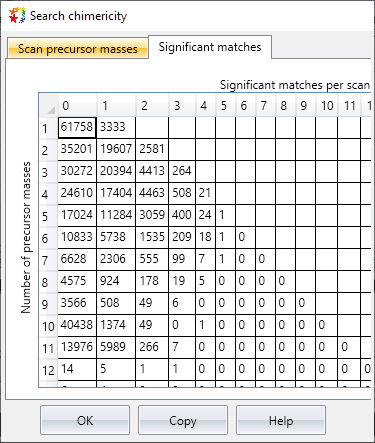
Distiller includes a new ‘chimericity’ report, which tabulates the count of detected and identified precursors per scan.
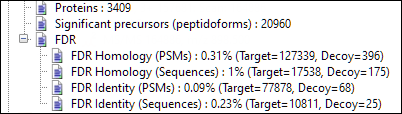
And the Search Header (Proteins tab) now displays the number of statistically significant precursor (peptidoform) identifications, a commonly used metric when comparing DIA software packages.
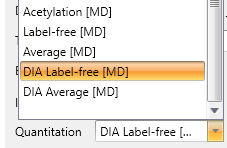
New: Quantify peptides and proteins from DIA data
Mascot Distiller implements MS1 quantitation for both DIA and DDA, and the full range of quantitation methods is supported (LFQ, SILAC, 18O, metabolic, etc.). Mascot Server 3.2 ships with two new LFQ protocols for Distiller, optimised for DIA:
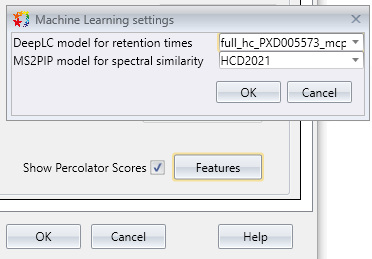
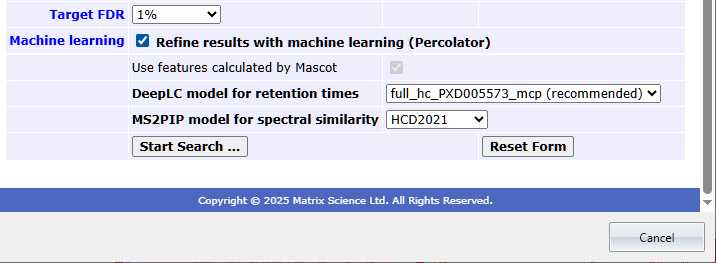
New: Boost identification and quantitation with machine learning [DDA, DIA]
Mascot Distiller now has a user interface for refining Mascot search results with machine learning. Tick a box to enable, then optionally choose a model for predicted retention times (DeepLC) and fragment intensities (MS2PIP).
Refining can also be selected in the embedded search form.
Machine learning is run by Mascot Server and the refined peptide identifications automatically imported into Distiller. Enabling machine learning can easily increase the number of quantified peptides by 25% while maintaining the same accuracy. Requires Mascot Server 3.1 or later.
New: Run all processing steps from the command line
Every processing step can now be run on the command line by calling MascotDistiller.exe: peak picking, search submission, quantitation and quantitation reports. The command-line arguments are documented in Help, Reference, Command line arguments.
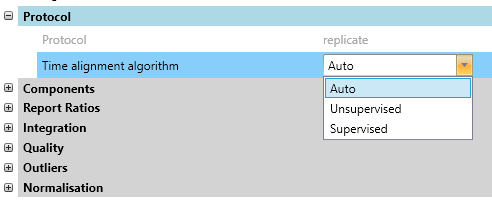
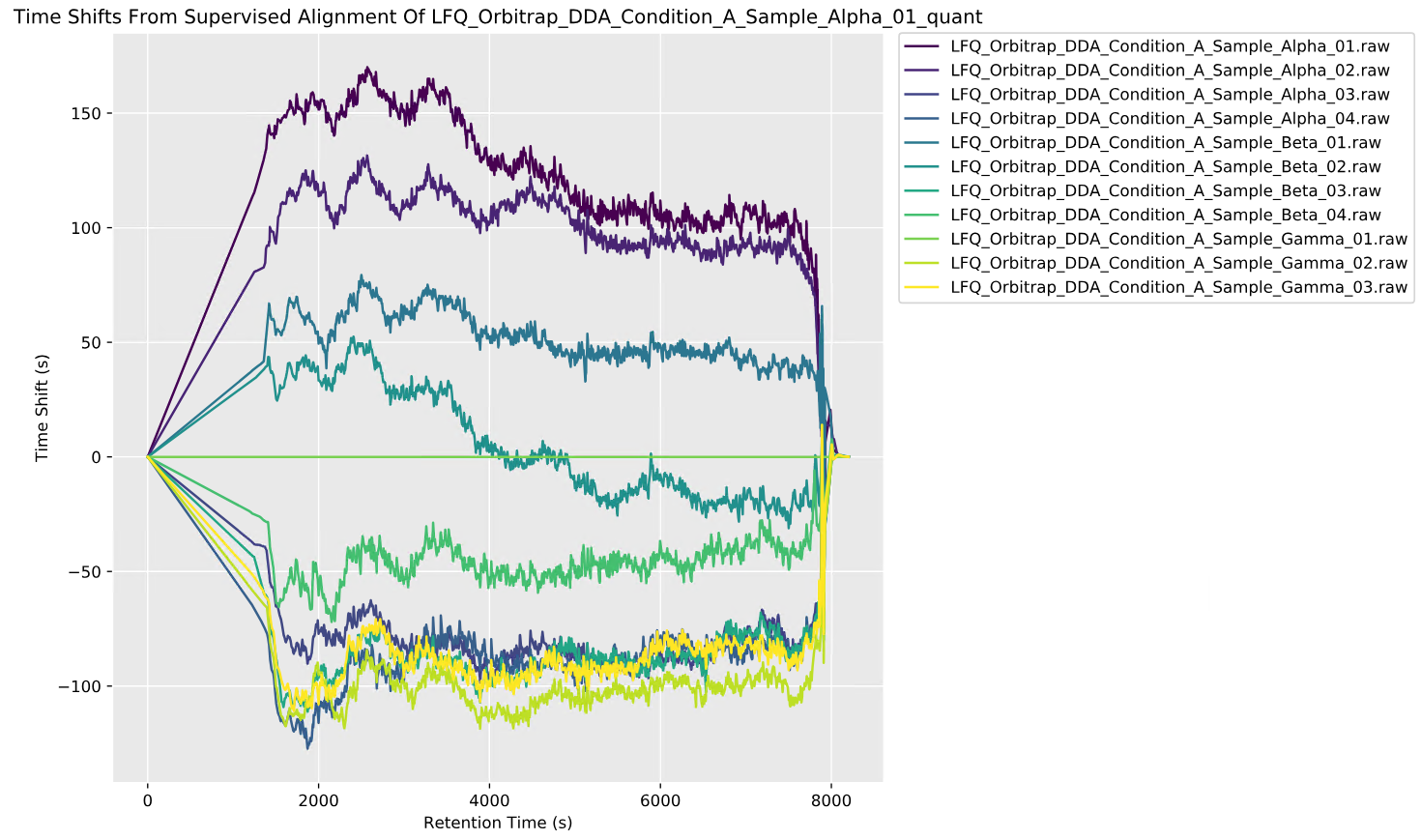
Improved: Supervised time alignment for Replicate quant [DDA, DIA]
Time alignment for label-free quantitation using the Replicate method now defaults to Auto, which selects between a supervised and unsupervised algorithm.
The previous algorithm is now called Unsupervised. The unsupervised algorithm uses precursor mass and XIC to align the runs, which allows aligning unidentified MS1 features. However, the unsupervised algorithm can take a long time to run.
The supervised algorithm uses high-confidence shared peptide matches as anchor points between runs, which makes time alignment both faster and more accurate. If a peptide is not detected in all runs, then for alignment purposes, Distiller now imputes a retention time using K-nearest neighbours or MissForest clustering.
A new time alignment report plots the non-linear correction function for each sample:
Improved: Replicate and Average quantitation on any project
In previous versions, for label-free quantitation Mascot Distiller required selecting a quantitation method in the Mascot search parameters. If you forgot to select an LFQ method, you had to repeat the database search for no good reason.
Mascot Distiller 3.0 now allows Replicate and Average quantitation on any project. The only exclusions are error tolerant searches and intact crosslink searches.
We have also added a new Top 3 protein intensity report, which calculates the Average (top-3) protein intensity for all sample components and generates a CSV export file.
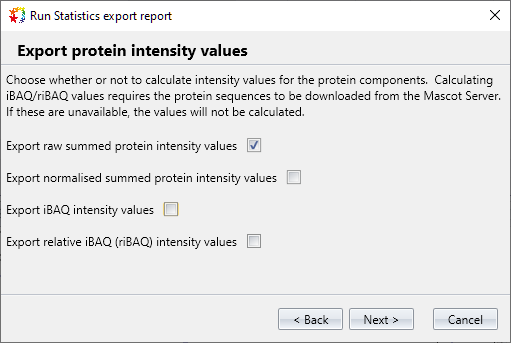
Improved: Stats export for label-free protein quantitation with iBAQ and riBAQ
The statistics export saves the protein quantitation data in CSV format, suitable for many stats packages like Perseus and R. The export has been extended, and you can now choose to export raw summed protein intensity, normalised summed protein intensity, iBAQ and relative iBAQ (riBAQ).
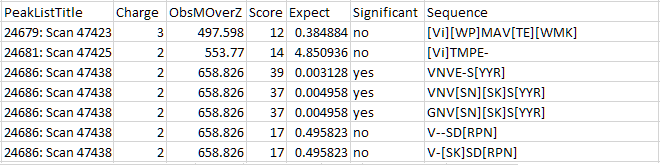
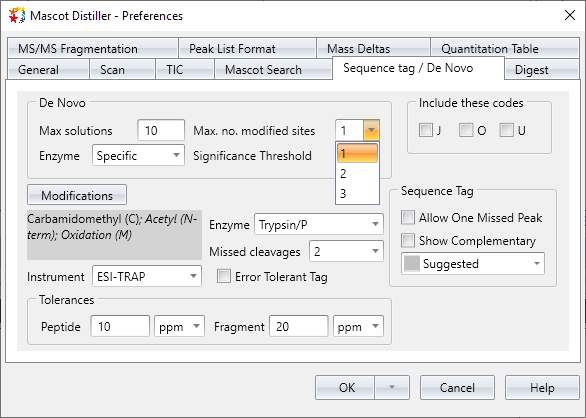
Improved: De novo statistics and variable mod handling [DDA]
De novo peptide identification now provides a statistical confidence estimate: the familiar expect value. After running a de novo search of a TIC range or all spectra, save the results as CSV (File, Export All Denovo Solutions), which has the new Expect value column. The default significance threshold is 0.05 and can be changed in Distiller preferences.
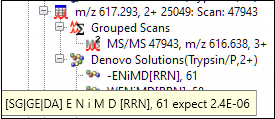
The expect value is also shown in the tooltip in the peak list tree.
You can now select more than one variable modification per de novo solution. The default is one and the limit can be increased to up to three variable modifications.
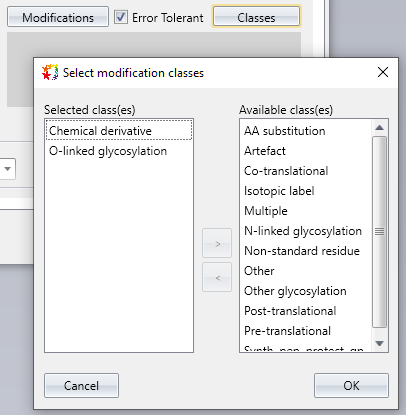
Improved: Select modification classifications for error tolerant search
Mascot Server 3.0 introduced a search parameter for modification classifications for error tolerant (ET) search. Mascot Distiller now has a user interface for selecting these.
The ET search is a two-pass search. In the second pass, Mascot Server tries to identify unmatched spectra by iterating over unexpected modifications in all the selected modification classes.
Performance improvements
Mascot Distiller now downloads results from Mascot Server using the new file format (MSR), provided the server is Mascot Server 3.0 or later. The MSR format is an SQLite database, which provides much improved read performance, especially with large searches and DIA searches.
If your processor has more than 64 logical cores (e.g. 32 physical cores with hyperthreading), then Windows may split the logical cores into processor groups. Previous versions of Distiller were limited to the first processor group. On Windows 11/Server 2022 and later, Distiller is now aware of processor groups and will utilise all logical cores in all processor groups.
Distiller now opens subprojects of an existing multifile project in parallel.
A new command-line parameter has been added for setting processor affinity. This is relevant on heterogenous processor architectures, such as Intel Alder Lake/Raptor Lake processors with ‘P’ (performance) and ‘E’ (efficiency) cores.
The user interface responsiveness has been improved by making more methods asynchronous.
When re-quantitation is triggered, the saved time alignment data is now reused to avoid triggering re-alignment.
Changes to defaults
default.ThermoXcalibur.opt has been reviewed to work better with profile data.
A new setting has been added to override.ini to control autosave for multifile projects. Autosaving is enabled by default. If Distiller crashes during a processing step, results from the previous steps are not lost.
For single peak picking (reporter ion region), MS/MS peak picking now has an option to take the mass range from the reporter ion masses defined in the quantitation method.
There is a new option to scale the TIC Y axis to give a reasonable view when you have XICs present.
A new Quantitation Settings menu item has been added to the analysis menu. Quantitation settings are also available via Tools, Format Options, Quantitation Options tab.
The minimum peak signal-to-noise ratio has been reduced to 0.001 in order to support single-cell datasets with reporter ions.
Removal of obsolete functionality
Mascot Distiller 3.0 requires Windows 10 or later. Support for Windows 8.1 and earlier, and Server 2012 and earlier, has been dropped.
Distiller now requires Microsoft .NET 4.7 or later.
Mascot Distiller no longer requires Internet Explorer. The embedded component of Microsoft Edge is now used for rendering the Mascot search form.
Other improvements
The release also fixes other minor issues. In total, over 170 change requests and bugs are addressed in this release. The full list is included in the release notes.