The promise of spectrum-centric DIA
We’re working on a universal spectrum-centric solution for the analysis of DIA data, called Mascot DIA. A preview was presented at our ASMS 2025 Breakfast Meeting, whose slides are free to download. But what is spectrum-centric searching and what is it meant to solve?
Complementary approaches
“Peptide-centric” and “spectrum-centric” analysis are broad technical terms that are mainly of interest to search engine developers. They are complementary ways of analysing bottom-up LC-MS/MS data. Here’s a simple but useful definition:
Peptide-centric
The goal of peptide-centric analysis is to find evidence for a theoretical precursor in the LC-MS/MS run.
Most peptide-centric tools start from a chromatogram library, spectral library or FASTA file, and make a list of theoretical precursors (peptides) at each time point in the elution. This relies on predicted retention times from a machine learning model or retention times from DDA runs. For each potentially detectable precursor at that time point, peptide-centric tools follow the elution of its fragment peaks in the MS/MS scans. If a sufficient number of highly correlated fragment traces are found, the precursor is considered identified.
Spectrum-centric
The goal of spectrum-centric analysis is to explain as many peaks in the MS/MS spectrum as possible.
Most spectrum-centric tools start from a FASTA file, and make a list of theoretical precursors (peptides) by digesting the protein sequence database. The list is filtered by precursor mass: only peptides within tolerance of the observed precursor mass are selected for in silico fragmentation. The fragments from each candidate are matched to the spectrum. If a sufficient number of theoretical fragment peaks match the observed peaks, the precursor is considered identified.
Characteristics of spectrum-centric analysis
Both approaches will ultimately produce a list of identified peptides, and a protein is then identified if a peptide sequence unique to the protein is identified.
Is spectrum-centric better than peptide-centric? It depends on what you need to identify. Here are some of the characteristics of spectrum-centric analysis.
1. Identification requires accurate precursor mass. The list of candidate peptides is based on precursor mass filtering, so spectrum-centric identification requires evidence that the precursor ion is detectable. Traditionally, in DDA analysis, precursor mass is determined from the MS1 (survey) scan. In DIA analysis, precursor masses can also be deduced from the MS/MS spectrum. Conversely, peptide-centric identification is possible even without precursor mass evidence, because it relies on tracing the elution of fragment peaks.
2. A precursor with few product ions won’t be identified by mistake. Spectrum-centric identification will never “detect” a peptide based on just 3-5 fragments. Confident identification requires a good run of b/y ions and decent sequence coverage, which means spectrum-centric identifications are inherently more reliable. On the other hand, peptide-centric analysis is better at identifying peptides in the middle ground. For example, a precursor that produces a handful of unique fragment masses that can be traced in sequential MS/MS scans is detectable with peptide-centric analysis.
3. All the information in the MS/MS spectrum is used. A spectrum-centric peptide identification must account for not just b/y ions, but also for fragment neutral loss and secondary ion series (like y-H2O). Many variable modifications like phosphorylation have signature neutral loss peaks, and different modification permutations may differ by just 1 fragment peak. This is very difficult to handle with a peptide-centric approach. Spectrum-centric analysis is the gold standard for modification site localisation.
4. Spectrum-centric analysis can be independent of retention time predictions. LC separation (and/or ion mobility separation) is invaluable with DIA, because it reduces the complexity of MS/MS spectra. However, when the basic analysis unit is the MS/MS spectrum, the thousands to millions of spectra in an LC-MS/MS run are analysed independently. You don’t need to acquire a chromatogram library; it doesn’t matter which columns you use or if your peptides come out in a different order than predicted by a model.
5. Spectrum-centric analysis can be independent of fragment intensity predictions. In particular, Mascot probability-based scoring does not use predicted fragment intensities. The independence has direct consequences: analyse DIA spectra from instruments that don’t have trained machine learning models, and identify precursors for which spectral libraries and ML predictions aren’t available.
What Mascot DIA aims to solve
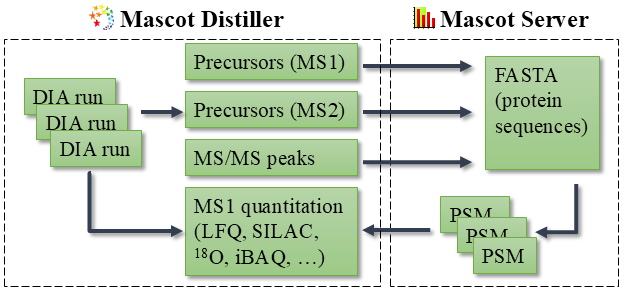
Mascot DIA is a spectrum-centric solution. Mascot Distiller processes DIA runs into precursor masses and peak lists, which are submitted to Mascot Server for database searching. The results are imported into Distiller, which runs quantitation.
The spectrum-centric approach has several application areas where peptide-centric DIA struggles: post-translational modifications; identifying endogenous peptides; semi-specific cleavage and enzymes other than trypsin; samples from any species (not just human or yeast); and MS1 quantitation methods based on a precursor mass shift like SILAC, 18O and metabolic labeling (not just LFQ).
We will provide more details about precursor detection from MS/MS scans and noise-resistant probabilistic scoring in future blog posts!
Keywords: DIA, modification, site analysis