Trying to illuminate proteomics ‘dark matter’
The May 2017 issue of Nature Methods has a paper from Alexey Nesvizhskii’s group at U. Michigan describing a new open database search program called MSFragger. Strikingly, they also observed the two highly abundant but unidentified mass deltas reported in Steven Gygi’s 2015 mass tolerant paper: 301.9864 and 249.9803.
The challenges of open searching were discussed in an earlier blog article. In particular, artefactual modifications due to mis-assigned precursors and lower PSM scores because only unmodified fragments are matched. These issues are discussed in the MSFragger paper, but are still to be solved.
A significant effort has been made in the MSFragger paper to assign as many of the observed mass deltas as possible. The authors find that several of the unassigned deltas appear to be labile modifications, which shift the precursor mass but have little or no effect on the MS/MS spectrum. This is perfectly possible, just think of sulfation or O-linked glycans under CID conditions. Of course, it would also be consistent with an artefact that systematically shifted the precursor mass, although it is hard to imagine a mechanism.
It is frustrating that apparently common and abundant mass deltas remain unidentified, so we want to try the power of crowd-sourcing. We have selected 20 abundant and unassigned mass deltas from Supplementary Table 3 of the MSFragger paper:
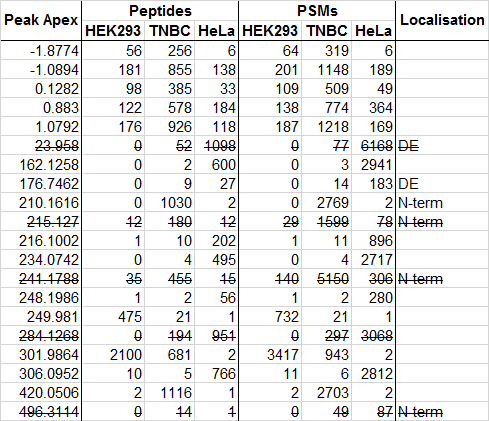
Data sets are
- HEK293: Pride project PXD001468
- TNBC: ProteomicsDB accession PRDB004167
- HeLa: Pride project PXD000612
For each of these deltas, we are offering a prize $100 (US Dollars) to the person who first reports a credible assignment. Credible means
- A possible chemical formula or reaction pathway, not just an elemental composition
- Chemistry must be consistent with the sample preparation and analysis as described in the associated paper
- Exotic elements or unnatural isotopes cannot be introduced without some reasonable explanation
- Mass accuracy appears to be excellent, so the assignment needs to be within 0.005 Da of the experimental delta
Competition rules
- To submit a proposed assignment, it must be entered as a comment to this article, one assignment per comment
- Assignments submitted in any other way are not valid
- You don’t have to use your real name, but you must use a genuine email address (which is not displayed) or we will have no way to make contact if you win
- One person can submit proposed assignments for as many deltas as they wish, and win multiple prizes
- An assignment that has already appeared in the literature is fine (please include the citation)
- Closing date 31 December 2017
- Matrix Science employees and their immediate families are not eligible to enter
- The judges decision is final
Credible assignments to date
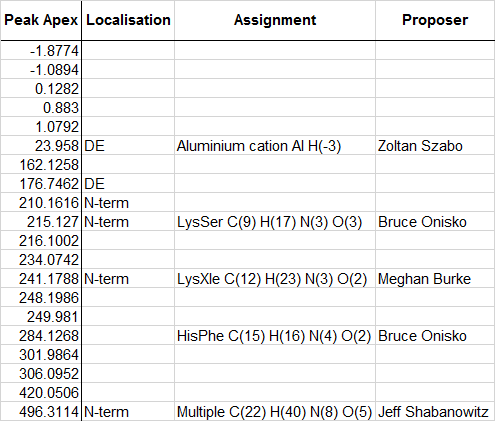
Keywords: dark matter, delta mass, error tolerant, mass tolerant, modification, open search, Unimod
23.958 Replacement of 3 protons by aluminum:
25.98689169 – 3 x 1,00794 = -23.95771863
Beacuse of localization at DE overall z<3 is also possible.
Sorry,
26.98153863 – 3 x 1,00794 = 23.95771863
Dear Szabo,
I am interesting about your finding “23.958 Replacement of 3 protons by aluminum”
But I wonder how this modification is been forming.
1.Can it be formed in any amino acid of certain proteins?
2. And how is it existing steadily:aluminum is a form of Al3+ ? and which three protons is replaced. I guess H in the “-CO-NH-“ of the protein/peptide is the probably one,but how about three protons?
I do not wish to receive a prize for the assignment; however, in my experience the peak apex at 241.1788 Da is due to semi-tryptic peptide formation corresponding to the addition of Leu/Ile and Lys. The monoisotopic mass for this addition is 241.179027 Da, which has a mass difference of 0.000227 Da compared to the mass reported.
23.985: monoisotopic mass of magnesium, explained by adduct formation with aspartate and glutamate with magnesium. For structures of Mg-glutamate and Mg-aspartate see
pubchem compound 198473 and 16211203, resp. Complexation may be enhanced when D and E are adjacent or nearby in the sequence.
One explanation that this was found in HeLa and TNBCs but not HEK293 may be that HEK cells are usually harvested by EDTA, detaching cells but also chelating Mg2+. Another possibility is that EDTA was added during tryptic digestion, to enhance tryptic activity by chelating Mg2+. Details how cells were harvested, as well as buffer composition during digestion are not mentioned in the papers.
Regarding the answer for 23.958, why is the average mass for the Hydrogen atom being used? The mass of a Hydrogen is 1.007825 and the mass for a proton is 1.00728.
Right, only makes a difference in the 4th decimal place, but it should be the monoisotopic mass for H. Al H(-3) = 26.981538 – 3 x 1.007825 = 23.958063
Hi John,
Thanks, the answer still comes out ok, just wanted to be clear on the masses.
Also, any idea where the Aluminium is coming from in those 2 experiments? Jeff
Sorry for H mass, I have just copied data to post but made mistakes.
The Hela experimental procedure includes TiO2 phoshpho enrichment and SCX/SAX ion chromatography so I think some of the mass shifts dominant in that data-set may be related to some of those steps, including contamination with Aluminum. But that’s just an idea, needs to be confirmed (or confuted :-).
Data +215.127 Ntm; proposed answer +SerLys (or +LysSer); +C9H17N3O3=215.1269; error = 0.0001 Da.
mechanism semi-tryptic cleavage. This could be confirmed by examining the sequence of the protein that includes the “modified” peptide, or redoing the open mod search choosing semi-tryptic for the enzyme.
Data 284.1268; proposed answer +PheHis (or +HisPhe); +C15H16N4O2=284.1273; error = 0.0005 Da.
John,
Curious, one would have thought all of these mass deltas would have been checked for semi-tryptic or non-specific cleavages before being included in this list. Looking at the table, but without actually researching all of the data, it would also seem odd that there would be so many actual peptides sequences exhibiting these dipeptide mass shifts (215.127=KS;241.1788=KX;284.1268=HF).
Jeff
p.s. by this same logic, one can explain the mass delta of 496.3144 and anyone of the following amino acid residue combinations: GGXXR; AGXVR; XVQR; AAVVR; or XXNR; wgere X=Leu/Ile. All of which calculate out to 496.31215, ~+1.5 ppm.
Excellent, another one explained.
We just took twenty of the more abundant un-annotated deltas from the MSFragger paper. Hopefully, the fact that some of them are not too difficult will encourage people to work on the others. It’s 250 and 302 that I’m really curious about.
Some of these short peptide deltas could be fully tryptic peptides with 2 missed cleavages. The MSFragger paper says the index was built with 1 missed cleavage, so presume a peptide like K.SKLPKPVQDLIK.M would be matched as K.LPKPVQDLIK.M with an N-term mod of SK.
306.0952: A possible explanation is the addition of a disaccharide (2 hexose units) through Schiff-base formation to lysine or to the N-terminus (dehydrated form, -H2O). Known from “glycation reactions” e.g. addition of glucose to lysine. Although relatively unstable, this and “dehydrated” forms have been described e.g. in reference Barnaby, Omar S., “Characterization of glycation sites on human serum albumin using mass spectrometry” (2010). The same reference may give also a hint for the nature of the adduct 234.0742 (with a delta of 0.0003 Da). Carbohydrates could derive from cell growth media, or from brake-down products.
Disaccharide (2xHex) C12H22O11 = 342.1162;
Schiff-base with Lysine C12H20O10 = + 324.1056 (“glycosyl-lysine”;
Dehydrated “glycosyl-lysine” adduct (-H2O) C12H18O9 = + 306.0951 (delta of 0.0001 Da.
C12 H18 O9 is an excellent fit to the mass delta. But, looking at Supplementary Table 3 of the MSFragger paper, the counts of PSMs/peptides in the HeLa data for related glycation masses are:
78/22 Hex (162.0528)
ND Hex-H(2)O (144.0423)
ND Hex(2) (324.1057)
2812/766 Hex(2)-H(2)O (306.0951)
ND Hex(2)-H(4)O(2) (288.0845)
(ND means not in top 500). Is this a likely pattern?
Is the ‘hint’ as to 234.0742 carboxyethyl+Hex? Again, a very good fit for the mass, but is this a believable structure when the counts are as follows?
40/16 carboxymethyl (58.0055)
ND carboxyethyl (72.0211)
78/22 Hex (162.0528)
ND carboxymethyl+Hex (220.0583)
2717/495 carboxyethyl+Hex (234.0740)
ND Hex(2) (324.1057)
ND carboxymethyl+Hex(2) (382.1111)
ND carboxyethyl+Hex(2) (396.1268)
Yes, fully agree that would be really unlikely. Except there is an unknown source for a disaccharide originating from (an unusual) medium or the sample prep. But again, rather unlikely looking to their paper and that Hex(2) is ND (not sure whether to expect the di-hydrated form). The hexose modifications proposed by Bruce below, sound like an interesting alternative for both, 306 and 234.
306.0952: I propose that the modification has composition C12H18O9. The hydrated modification would have composition C12H20O10. This composition fits for the ester of 3-hydroxy-3-methylglutarate and glucose. The 6-O-acetyl-, and 6-O-malonyl-glucose derivatives are known (see for example ChEBI entry 59475). I propose a protein modification by glucose (probably O-linked as usual to Ser or Thr) and then esterification of the 6-hydroxyl of glucose with 3-hydroxy-3-methylglutarate. If correct, this would be a modification (C6H8O4 or 144.042) of a glycosyl modification. The C6H8O4 modification is in UniMod already (two different isomers), but I like esterification as described since 3-hydroxy-3-methylglutaryl coenzyme A is such an abundant intermediate of metabolism and a substrate of many enzymes. The structure could be confirmed by synthesis. (No need for prize if correct, thanks.)
234.0742: I propose that the modification has composition C9H14O7 (234.0739). The hydrated modification would have composition C9H16O8. This composition fits for the ester of lactic acid and a hexose. I propose a protein modification by glucose (probably O-linked as usual to Ser or Thr) and then esterification of the 6-hydroxyl of glucose with lactic acid. If correct, this would be a modification (C3H4O2 or 72.0211) of a glycosyl modification. The C3H4O2 modification is in UniMod already (four different isomers), but I like esterification as described since lactic acid is such an abundant intermediate of metabolism and a substrate of many enzymes. The structure could be confirmed by synthesis. (No need for prize if correct, thanks.)