Disulfide bond characterisation
Support for matching crosslinked peptides is a new feature in Mascot Server 2.7. The following linkage types can be detected:
- intralinks – the linked peptides are from the same protein
- interlinks – the linked peptides are from two different proteins
- looplinks – connecting two amino acids within a single peptide
- monolinks – a linker with one end attached and the other free
The variety and complexity of crosslinking experiments means that analysis software sometimes needs a large number of parameters to define how to process the data. We have chosen a similar approach to that used for quantitation, and placed these new parameters into named crosslinking methods, one of which can be selected for a search.
The most widely encountered example of a natural crosslink is the disulfide bond, found in nearly one third of eukaryotic proteins. In most experiments, the location of these bonds is of no interest, so they are cleaved with a reducing agent, prior to analysis, and the cysteines alkylated to prevent bridge re-formation. Nevertheless, disulfide bonds are crucial to protein folding and tertiary structure stability. A paper from the Brodbelt group at University of Texas at Austin explored the use of 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry to interrogate disulfide bond patterns in lysozyme and serotransferrin. A particular difficulty occurs when two disulfides interlink, so that one must be broken to characterise the other, as in chicken lysozyme:
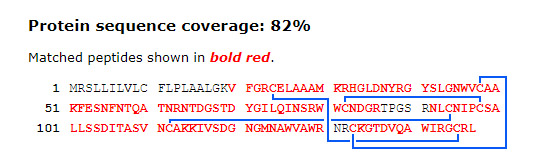
The study used a time course experiment with TCEP reduction and NEM alkylation to help elucidate such cases. We downloaded a raw file from ProteomeXchange for chicken lysozyme (Lysozyme_2p2mJ_1minute.raw) and processed it in Mascot Distiller 2.7. Fragment masses were de-charged to 1+ because fragments that include the linked peptide are likely to have charge states higher than 2+, which means they will not be matched as m/z values. SwissProt was searched using Mascot Server 2.7, with these settings:
Crosslinking : Disulfide bridge in Lysozyme Enzyme : Trypsin/P Variable modifications : Nethylmaleimide (C), Oxidation (M) Peptide mass tolerance : ± 20 ppm Fragment mass tolerance : ± 20 ppm Max missed cleavages : 2 Instrument type : ESI-TRAP
The crosslinking method was unusually simple, specifying the Unimod name of the linker, the protein accession, and specifying that we were looking for intralinks and looplinks. You can view the settings by clicking on the method name in the results summary header. Reference information can be found here.
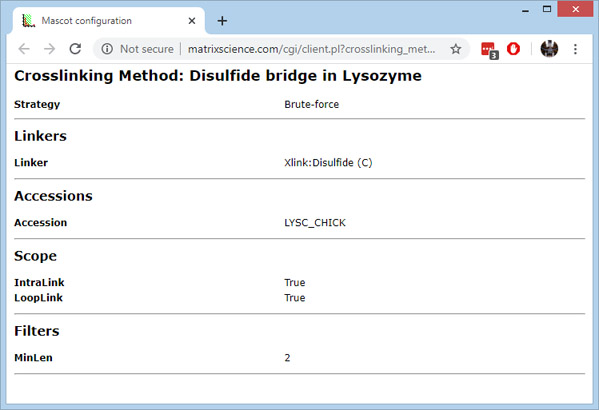
The MinLen setting enables matches to be reported for crosslinked pairs where the length of one peptide is just two residues. In the absence of this setting, the default would be the general peptide length threshold of 7.
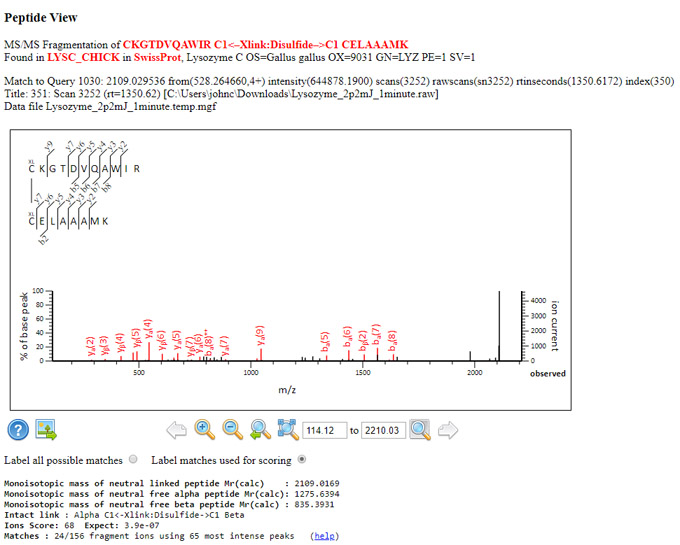
The result report is here. Four disulfide bridges are annotated in the SwissProt entry for LYSC_CHICK. All were detected, although one was only a weak match:
- FT DISULFID 24 145: Query 550, score 24 (weak match)
- FT DISULFID 48 133: Query 1118, score 101
- FT DISULFID 82 98: Query 1495, score 91 (bridge 4 reduced and alkylated)
- FT DISULFID 94 112: Query 1101, score 65 (bridge 3 reduced and alkylated)
The raw file represents the first time point, where some spectra suggest there may be a degree of scrambling. For example, the looplink in the match to query 1101 is clearly between 94 and 98, rather than the expected 94 and 112, because we see extensive fragmentation between residues 100 and 112, which would not be possible if the link spanned this region. Another discrepancy would be the match for query 1030, which has an unambiguous link between C24 and C133 rather than C24 and C145.
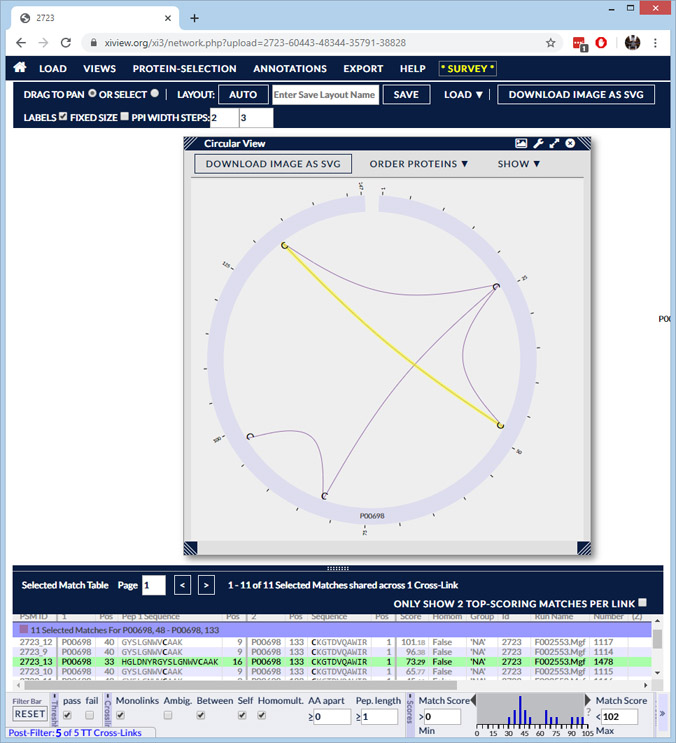
Search results for crosslinked peptides can be exported to xiVIEW for visualisation. This is very useful for protein-protein interaction experiments, where relationships between proteins can be explored. For a disulfide bond experiment, it can be helpful to display the links as a chord diagram:
Keywords: crosslinking, export, Mascot Distiller, modification