Not Better Together: Processing and searching combined CID+ETD datasets with Mascot Distiller 2.6
Many modern mass spectrometers will produce raw datasets containing more than one type of scan data. For example, a common experiment using a Thermo Orbitrap instrument is to select the most abundant precursor ions and either independently fragment them using both ETD and CID fragmentation, or use a decision tree to select different fragmentation methods depending on, for example, the charge state [e.g. Frese, C. K et al., J. Proteome Res. 10, 2377–2388]. These different fragmentation methods will usually produce different ions series for the same fragmented peptide. A more detailed overview of the different ions you can expect from peptide fragmentation using different methods can be found on this help page.
The set of ions series to be considered in a Mascot search are specified by selecting the Instrument type from the drop down list on the search form. If an ions series is not part of the selected instrument definition, then it will not be considered by the search engine, no matter how intense the peaks are. Therefore, it is important that your instrument definitions contain all of the MS/MS ions series that you expect to see for the particular instrument type. However, you should not include all possible ions series just in case. Doing that will increase the search space and decrease the search specificity, meaning that while the score for the correct match is unchanged, the score distribution for random matches is increased.
If you are searching a peaklist file which contains MS/MS peaklists derived from spectra generated using different fragmentation methods, one option is to create a hybrid instrument definition which contains the expected ions series from all of the fragmentation methods represented in the file. An example of this is the combined CID+ETD instrument definition that we’ve been shipping as part of the standard Mascot installation since Mascot 2.2:

As you can see, this instrument definition contains the combined ions series from the CID and ETD instruments. If your Mascot server does not contain this instrument definition, then you can find details on how to add it in chapter 6 of the Mascot Setup & Installation Manual.
The disadvantage of this approach is that we’re always including non-significant ions series in the Mascot search for each MS/MS peaklist that is searched – for example, we’ll be including b-series ions in searches of ETD derived peaklists and c-series ions in searches of CID derived peaklists. As described earlier, this will increase our search space and decrease the search specificity. In theory, you could get better results if you could search each peaklist type using the correct instrument definition.
Since Mascot 2.2, the Mascot Generic Format for peaklists has supported setting the instrument type at the level of each individual MS/MS peaklist. In Mascot Distiller 2.6, on the ‘Peak List Format’ tab under preferences, you’ll find an additional option which allows you to automatically set the INSTRUMENT parameter in the exported peak lists of a selected scan type. For these spectra, when the peaklist is generated, the INSTRUMENT parameter value will override the option selected on the Mascot search form:
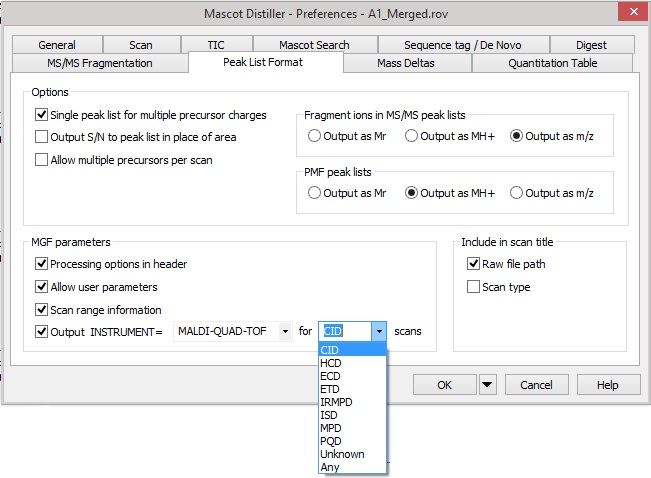
To examine the improvement in search results from using the separate instrument search parameters output by Mascot Distiller 2.6, we took one of the technical replicates from a dataset in the EBI’s PRIDE archive which contained both CID and ETD spectra in the same raw data files (PXD000293). The authors used a decision tree to determine whether precursor ions were fragmented by CID or ETD [de Graaf EL et al., Mol Cell Proteomics 13(9), 2426-2434]. The raw data files were reprocessed in Mascot Distiller and we then carried out two separate Mascot searches – one using the combined CID+ETD instrument, and one using the correct, separate instrument definitions. All other search parameters were identical between the searches and the significance threshold adjusted to give a 1% peptide false discovery rate once completed:
| Search | Target peptide matches at 1% FDR |
|---|---|
| Combined CID+ETD instrument | 33174 |
| Separate instrument definitions | 37412 |
As you can see, using the separate instrument definitions output by Mascot Distiller 2.6 yields a 12.8% increase in the number of significant peptide matches identified at a peptide FDR of 1% for this dataset. In addition to that, the search runs faster because the search space is smaller:
| Search | Search duration (minutes) |
|---|---|
| Combined CID+ETD instrument | 114 |
| Separate instrument definitions | 90 |
So, while using the combined instrument definition is giving acceptable results, we are seeing significant improvements in both the results and the speed of the search by separating out the instrument definitions for the two different types of spectra, a process which is made trivial in Mascot Distiller 2.6.
Keywords: fragmentation, ion series, Mascot Distiller