Step away from the iodoacetamide
In our July newsletter, we featured a paper from Torsten Müller and Dominic Winter, University of Bonn, concerning alkylation artefacts. Some of their findings were quite shocking. For example, differences of more than 9 fold in numbers of identified methionine-containing peptides for in-gel digested samples between iodine- and non-iodine-containing alkylation reagents.
This is important because a glance at the literature or any data repository will show that iodoacetamide is by far the most widely used alkylation reagent, as is confirmed by supplementary Figure 1 in the Bonn paper:
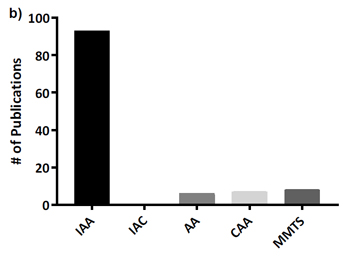
(Note that Müller and Winter use IAA as the abbreviation for iodoacetamide and IAC for iodoacetic acid, rather than IAM and IAA.)
The propensity of iodoacetamide for over-alkylation was highlighted back in 2001 by Boja and Fales, who commented "In examining a series of other peptides alkylated with IAM [when the IAM was allowed to remain during the digestion], we have found N-alkylation to be the rule rather than the exception". For an example of this, look at these search results from the recent blog article about trypsin artefacts. There are more PSMs with Carbamidomethyl (N-term) than with Carbamidomethyl (C).
In many searches, large numbers of PSMs are missed because only Carbamidomethyl (C) is selected for the search. Even if the investigator is aware of over-alkylation, and selects additional specificities, results take a double hit: (i) Additional variable modifications increase the search space so that sensitivity is reduced and searches take longer, (ii) Signal to noise suffers because the signal for any particular peptide is spread over multiple precursors. When it comes to quantitation, the second effect can be a major source of error.
Boja and Fales did not observe alkylated Methionine, even though this reaction is well known. This is explained by Müller and Winter, who found near quantitative, in-source loss of the side chain, making the net delta in the precursor mass -48 Da. The Unimod name for this modification is Dethiomethyl.
There are marked differences between the solution data and the in-gel digest data. For example, here are the results of our error tolerant searches on the DTT + iodoacetamide data downloaded from the associated Pride project PXD005183: solution (InSolution_DI_Rep1_pH*_RAW.raw) and in-gel (InGel_DI_Rep*_RAW.raw). Carbamidomethyl (M), which was missing from Unimod, was added with neutral losses of 0 and 105 Da. To clarify the modification counts, addition of Gly to K, S, and T, which has an identical composition to carbamidomethylation, was excluded from the error tolerant search.
The two searches have similar numbers of PSMs. Expand the modification statistics sections of the headers to see counts of modifications. The in-gel data has very high levels of non-cysteine alkylation. (Note that Ala->Gln and Gly->Asn are compositionally identical to carbamidomethyl and Val->Arg is near isobaric.) The count of Carbamidomethyl (M), where the side chain is lost on MS/MS fragmentation, is just 43 compared with 679 for Dethiomethyl (M), which is in-source fragmentation. In contrast, the solution data has little +57 except on Cysteine, but it does have high levels of +12 (Thiazolidine, listed in Unimod as a formaldehyde adduct). Is it possible that this is another degradation product of alkylated residues?To quote once more from Boja and Fales "Generally, when these alkylation experiments are run on macroquantities of intact proteins, one can carefully adjust the amount of IAM and reductant to avoid overalkylation. However, when it is a matter of running such reactions on a protein band isolated from a PAGE gel, such material balance is difficult to achieve and no recipes to avoid this problem seem suited for all purposes."
Given that there are alternative alkylation reagents that are far more specific to Cys, such as acrylamide, Müller and Winter make a strong case for their advice: "In general, the iodine-containing alkylation reagents (IAA and IAC) resulted in the worst identification rates irrespective of the digestion condition, or reducing reagent, and should therefore be avoided."
Keywords: alkylation, artefact, carbamidomethyl, error tolerant, iodine, iodoacetamide
And N-terminal Carbamidomethyl Cys will often abstract NH3. Then, a modification “loss of ammonia at N-terminal Cys” can be used.
//Jonas Nilsson, University of Gothenburg
A recent paper from the University of Sydney shows that 2-chloroacetamide is not a great choice because it increases levels of oxidation, especially for Met – http://dx.doi.org/10.1021/acs.jproteome.7b00022