Sequence database setup: Contaminants
|
Overview
If you search a single organism database, its usually a good idea to include sequences for common contaminants, such as keratins, BSA, and trypsin.
Two groups make their collections available for download. The Max Planck Institute of Biochemistry, Martinsried, maintains a file of approximately 250 proteins selected from various sources. The Global Proteome Machine Organization common Repository of Adventitious Proteins contains some 112 proteins selected from Swiss-Prot. (Numbers as of October 2011).
In Mascot 2.3 and later, you can simply select the contaminants database in the search form, along with the target database. For Mascot 2.2 and earlier, you need to append the contaminant sequences to the end of the target database fasta file. This can be complicated by the requirement to have a uniform syntax for all the title lines. One database may have Swiss-Prot style accessions and the other NCBI-style accessions. If so, you either have to find a parse rule that works with both or modify the title lines of one database using a script or text editor. If both target and contaminants databases have accessions drawn from the same pool, remember to watch for duplicates. It may be safer to add a prefix to the accessions of the contaminants entries so as to avoid possible collisions.
Download
https://lotus1.gwdg.de/mpg/mmbc/maxquant_input.nsf/7994124a4298328fc125748d0048fee2/$FILE/contaminants.fasta
for contaminants from MPI
http://ftp.thegpm.org/fasta/cRAP/crap.fasta
for cRAP from GPM
Taxonomy
Taxonomy is not appropriate. You want to include all contaminants in every search.
Parse Rules
Fasta title lines in the MPI collection vary according to the source database. Use standard rule 4 for the accession and standard rule 5 for the description.
Fasta title lines in the GPM collection contain SwissProt ID and no description. Use standard rule 4 for both accession and description.
Configuration (Mascot 2.3 and earlier)
The MPI collection was downloaded to C:\inetpub\mascot\sequence\contaminants\current, decompressed using gzip, and renamed to contaminants_20100513.fasta.
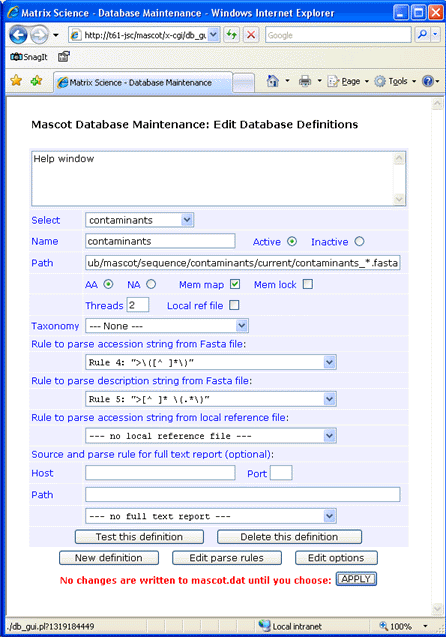
The GPM collection was downloaded to C:\inetpub\mascot\sequence\cRAP\current, and renamed to cRAP_20100324.fasta.
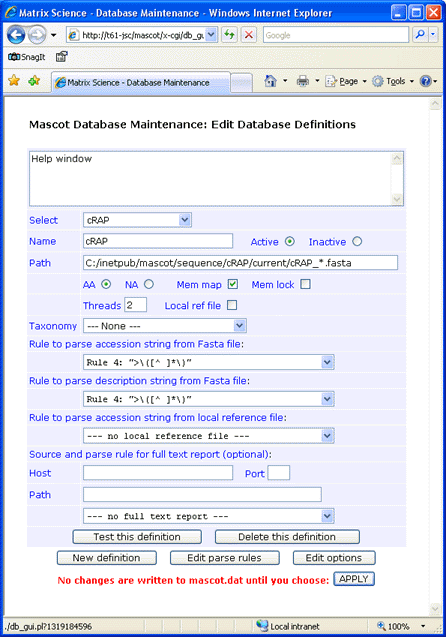
Always test a new definition before applying the changes to mascot.dat.