Sequence database setup: Manual configuration and updating
This page describes the procedure for downloading and configuring a sequence database without using the browser-based Database Manager. This information may be useful if you need to integrate the databases available for searching in Mascot with some external database management system. Otherwise, Database Manager is a much easier and more reliable option.
1. Choose a name for the Database
Give the database a short, descriptive name. This is the name that will appear in the drop-down list in the search form, so don’t write an essay. Note that database names are case sensitive. Allowed characters are alphanumerics and _.-$%()[]
2. Create a local directory structure
The recommended arrangement is to have a dedicated directory for each database. Within this directory are three sub-directories. The incoming directory provides a workspace for downloading and processing a new database file. The current directory contains the active database, and this is where Mascot Monitor creates the compressed files that will be memory mapped. The old directory is where the immediate past database files are archived, just in case.
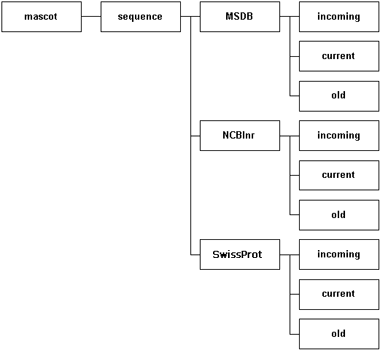
- Giving the database and the directory the same name is usually a good idea, but is not a requirement.
- There is no requirement for all the database directories to be placed in the mascot/sequence directory.
- Under Windows, path and file names are not case sensitive, but it is safer to treat them as if they were.
- Under Unix, links provide great flexibility, and the files or directories for a given database can be located wherever convenient. If the Fasta file is actually a link, then Mascot will create the compressed files in the directory containing the link, not in the target directory containing the Fasta file. If you want the compressed files to be on a remote drive, you can do this by making a link at the directory level. However, ensure that the network bandwidth is sufficient, and that the operating system supports memory mapping of NFS mounted files.
- Mascot does not support Windows UNC paths.
3. Download the database files
Download at least one release of the database manually, so as to verify the filenames and URLs. If the database is not described in one of the Mascot help pages, make careful notes of which files are required, where they come from, and any processing that is required.
Mascot can search any Fasta format sequence database. The Fasta format is extremely simple. Each entry consists of a one line title followed by one or more lines containing the sequence data in 1 letter code. Fasta databases can contain either amino acid sequences or nucleic acid sequences, but not a mixture. Nucleic acid databases are translated on the fly by Mascot in all six reading frames.
The Fasta title line begins with a "greater than" character, followed by some mixture of accession string(s) and description. Apart from the use of the "greater than" character, the precise syntax of the title line is not defined. The title line is delimited from the sequence that follows by a platform dependent new line character. Line lengths vary between databases; anything from 60 characters to a thousand or more. Mascot can handle lines up to 50,000 characters long. The end of a sequence is indicated when the following line is either a new title line or the end of the file.
The following characters are not allowed in a protein accession string:
- comma (,)
- double quote (")
- ASCII control characters (0×00-0x1f)
- Characters outside US-ASCII
Some databases come with a "reference" file, containing annotation text and cross-reference information. An example would be the SwissProt Dat file. Mascot can incorporate the full text for an entry into the Protein View report. If a full text file is not available for download, Mascot may be able to retrieve equivalent text from a remote HTTP server, as shown in the examples for NCBI and UniProt databases.
If database entries contain taxonomy information, Mascot can use this as a filter during a search. Many of the most popular databases from NCBI and UniProt include taxonomy. To determine taxonomy accurately, Mascot requires database specific supporting files. Details of these can be found in the help pages for the individual databases. Note that these supporting files have to be downloaded into the taxonomy directory, not into the sequence database directory. Also, some files need to be unpacked (using tar) as well as uncompressed.
4. Configure the database
Configuration information for sequence databases and associated files is stored in the general configuration file, mascot.dat. You can edit mascot.dat using any text editor, but first familiarise yourself with Chapter 6 in the Installation & Setup manual, Configuration and Log Files, which contains a comprehensive description of the configuration parameters.
In Mascot 2.4, Database Manager is the recommended tool to inspect and modify sequence database definitions. When Database Manager is used, it stores configuration information in a number of XML files and mascot.dat is dynamically re-written whenever a change is saved. This means that any direct edits to the affected sections of mascot.dat will be discarded.
5. Bring the Database on-line
Once you save a new definition to mascot.dat, Mascot Monitor will look to see if there is a Fasta file that matches the wild card path. If so, it will begin to compress the Fasta file (to minimise the memory requirements). If taxonomy has been defined for the database, Monitor will also create a taxonomy index.
Once this is complete, the new database is tested by automatically running a standard search. If this succeeds, the new database becomes available for general use.
6. Updates
In most cases, you’ll want to update the database files periodically. Whenever Mascot Monitor sees a new Fasta file in the current directory that matches the wildcard path, it will automatically swap to the new database, as described below. If the database has associated files, such as full-text or taxonomy or UniGene, make sure all these files are in place before renaming or moving the new Fasta file.
Databases can be updated as often as you wish, with no disruption to searches. Whenever Monitor sees a new Fasta file that matches the wild card path, the new database is compressed and tested. If errors are detected in the new database, the database exchange process is abandoned. Assuming the test is successful, all new searches are performed against the new database, while searches that are in progress against the old database are allowed to continue. Once the final search against the old database is complete, it is unmapped from memory and the files moved to the "old" directory. The new database is then memory mapped and the system becomes ready for the next update cycle.
Points to watch
- Remember the wild card in the database path
The wild card is important. First because it masks the time-stamp or version number. Second, because it allows the database to be updated without interrupting ongoing searches. Even if you don’t want to use a time-stamp or version number, you must still include a wild card. The filename should be like SwissProt_*.fasta or SwissProt*.fasta. If you specify the filename as SwissProt.*, then Mascot won’t be able to distinguish the Fasta file from the compressed files, with interesting results.
- Don’t use spaces or special characters in the database path
Spaces in paths may be legal in Windows, but they shouldn’t be. Apart from the wild card in the filename, only alphanumerics and the following characters are permitted in paths: _.-$%()[]. Although Windows uses back slashes in paths as the directory separator, all paths in mascot.dat must use forward slashes.
- Beware locking databases into memory
All databases should be memory mapped, because this makes access much faster. The benefits of locking a file in memory are not so clear. If you try to lock a database in memory and there isn’t enough room, the operation fails, and everything is OK. The real problem is when there is just enough RAM to lock the database, but very little left over for Mascot searches and other applications. Searches will be very slow, the disk will thrash, and eventually the system is likely to crash or hang.
- Don’t forget the taxonomy files
If database entries contain taxonomy information, Mascot can use this as a filter during a search. To determine taxonomy accurately, Mascot requires database specific supporting files, details of which can be found in the help pages for individual databases. These supporting files go in the taxonomy directory, not in the sequence database current directory.
- Check the statistics file after a new database has been compressed
In particular, verify that the number of entries is reasonable and that there are no entries reported as "too long". (If so, you need to increase the value of MaxSequenceLen in mascot.dat). If taxonomy is defined, look at the fraction of entries with no taxonomy. It is rare to have 100% success with taxonomy, but a failure rate greater than 1% is a cause for concern. Maybe the taxonomy files are out of date?
- Don’t stop Mascot Monitor when updating a database
There is no need to stop Mascot Monitor when updating a database, and doing so can actually cause problems. Just move the new files to the current directory, as described above, and Mascot will handle the exchange and move the old files to the old directory. If Monitor is stopped during database exchange, move the files for the old database from current to old before re-starting it.