Adding a custom AI/ML adapter to Mascot
Mascot Server 3.0 and later ship with MS2Rescore for predicting retention time and spectral similarity. What if you wanted to use a different AI/ML system for predictions? Mascot Server has an application programming interface (API) exactly for this purpose. The tutorial below includes a fully working Python script that you can use as a starting point for your own adapter.
What is an ML adapter?
Because predictive ML for proteomics is so new, every ML system has different configuration settings, different input and output formats and different runtime requirements. An ML adapter is a command-line application that translates data between Mascot Server and the target AI/ML system, and performs any other preparatory steps that are necessary for its use. This way, new integrations can be added without having to change Mascot Server — just add a new adapter.
The MS2Rescore integration is implemented using an ML adapter. However, an adapter doesn’t actually need to integrate with an AI/ML prediction system to be useful. It could compute metrics directly from the PSM data, or run a Python module or external executable, or connect to a database, or use HTTP to send PSM data and receive predictions from some service.
The full ML adapter protocol and data formats are described in chapter 13 of the Installation & Setup manual, which you can download from your local Mascot homepage.
Example adapter
The adapter can be written in Python, Perl, Java, C# or C++. For the purposes of this tutorial, we have developed example_ML_adapter.zip, which performs the following steps:
- Given a Mascot results file (--resfile), open it using Mascot Parser.
- Read and parse the query list file (--query_list), or exit if failure.
- Iterate over the query list. Compute peptide length for each PSM.
- Iterate over the query list again. Print the calculated values in the output file (--output).
When the adapter is activated, Mascot calls it with the search results and incorporates the ‘predicted’ feature (peptide length) into the Percolator feature set. Although the Mascot core feature set already has a feature called peptideLength, it doesn’t clash with the feature retrieved from the adapter. This is because Mascot prefixes the feature names with the name of the adapter.
An adapter can return any number of computed or predicted features. For simplicity, the example adapter returns just the one feature.
Running the script
To make sure Mascot can run the adapter, try running it from the command line:
- Install Python 3.6 or later if not already installed. This tutorial assumes Python is installed in C:\Python\Python311.
- Download and copy example_ML_adapter.zip to C:\inetpub\mascot\bin\ML_adapters\custom, assuming Mascot Server is installed in C:\inetpub\mascot.
- Download Mascot Parser from our website. Extract it in a suitable temporary directory.
- Copy msparser.py and _msparser.pyd from the python36_or_later directory to C:\inetpub\mascot\bin\ML_adapters\custom, adjacent to example_ML_adapter.py.
- Run the script to confirm all the components are in place:
C: cd \inetpub\mascot\bin C:\python\python311\python.exe ML_adapters\custom\example_ML_adapter.py --help
The script must be run from the mascot\bin directory, because it needs access to mascot\config\mascot.dat. However, the script itself does not need to be saved in mascot\bin. A convenient location is the bin\ML_adapters\custom directory.
The steps above have been tested on Windows. The steps on Linux are very similar; just use the Python version from your Linux distribution and change the filepaths accordingly.
Adding the ML adapter to the user interface
Mascot has a central configuration file for ML adapters, mascot\config\ML_adapters.toml. This file defines the adapter name, where it’s installed and what parameters it needs. Mascot uses ML_adapters.toml to create the dropdown menus in the search form and the Protein Family Summary report.
Follow the instructions in chapter 13 of the Installation & Setup manual to add the new adapter. Briefly, the following minimal configuration will work:
[example_ML_adapter] interpreter = "C:/python/python311/python.exe" program = "C:/inetpub/mascot/ML_adapters/custom/example_ML_adapter.py" visible_in_user_interface = true [[example_ML_adapter.parameters]] name = "enabled" title = "Example: example_ML_adapter.py" values = [ 1 ]
This assumes you have saved the script in C:\inetpub\mascot\ML_adapters\custom. Make sure you also copy _msparser.pyd and msparser.py to the same directory. The parameter value is not used by the example script, but at least one parameter must be defined to be able to activate the adapter in the user interface.
Open a target-decoy search on your local Mascot server. You should now have a new ML adapter option:
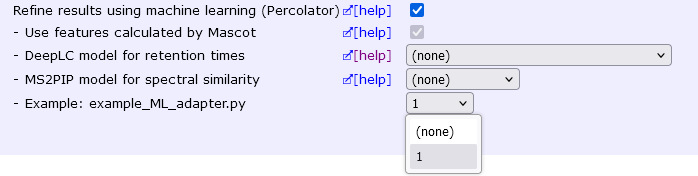
Choose ’1′ and Apply. If all goes well, you should now see the usual progress bars for machine learning, Percolator, and then caching the results.
Evaluating the computed feature in the ML quality report
In the Protein Family Summary report, open the machine learning quality report from the link provided:
Open the Rescoring Features tab. This tab displays a graph showing the Q-value ECDF AUC of each active feature. The larger the AUC, the better the feature is at discriminating between target and decoy PSMs.
Mascot groups core features separately from any features computed by ML adapters:
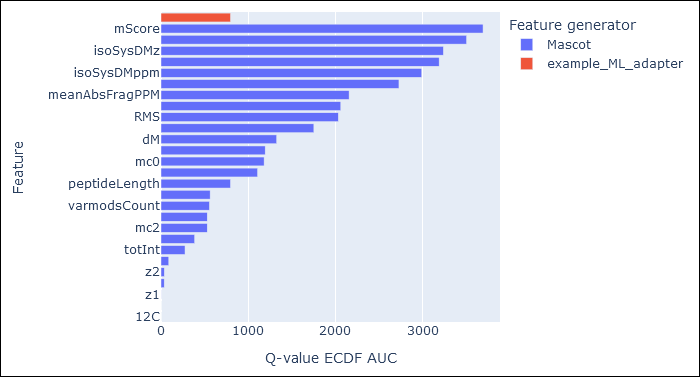
peptideLength is a core feature computed by Mascot. There is no issue with using the same feature name in some other adapter, because Mascot prefixes the feature name with the adapter name before combining with core features. In this case, peptideLength in core features is identical to peptideLength computed by example_ML_adapter.py, so they have identical AUC.
Where to go next?
The example script is a fully working starting point for your own AI/ML adapter. The adapter should be written in one of the programming languages supported by Mascot Parser: Python, Perl, Java, C# or C++. The Parser documentation is extensive, but for most adapters, you just need to be able to open the results file, load a peptide match and read the PSM properties (ms_peptide methods). The Mascot results file also contains the peak lists and full precursor information, available using ms_inputquery.
Keywords: machine learning, sysadmin, tutorial