Converting BLIB spectral libraries to MSP (and back again) for use with Mascot Server
Mascot Server can create and search NIST MSP formatted spectral libraries. However, BiblioSpec, which is part of the Skyline software, creates spectral libraries in the BLIB format which cannot be used. We have developed a script that can convert BLIB to MSP files or vice versa, MSP to BLIB files. The conversion script allows Mascot Server to search in silico libraries generated by Carafe or similar prediction software. Skyline can already read NIST (.MSP) files, but MSP libraries converted to BLIB format may be useful to other spectral library (SL) search engines and will allow one library to be used consistently across multiple platforms whatever the source.
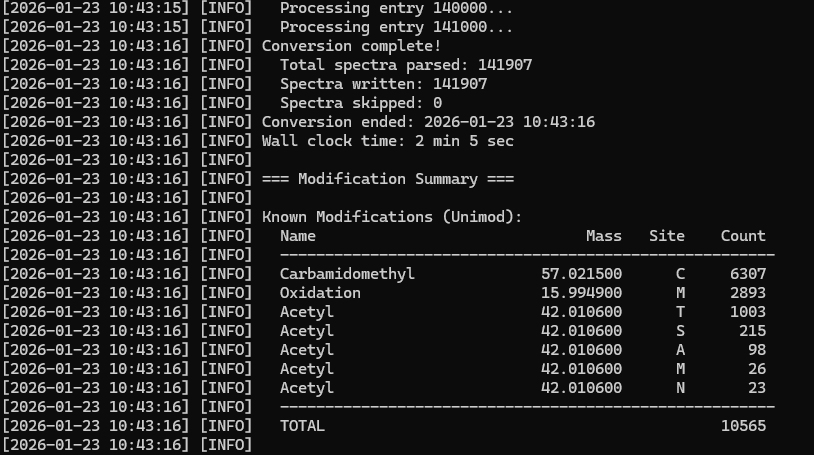
For testing purposes, I converted BLIB files from a Mycobacteria data set. The libraries ranged in size from fifty megabytes to two gigabyte in size. I also ran roundtrip tests where the library was converted from one format to another and back again with no differences between the original and regenerated libraries. The converter is available for download in our GitHub repository.
BLIB to MSP Spectral Library Converter installation and usage
The BLIB to MSP Spectral Library Converter is written in Perl and uses the Mascot Parser library. On Windows, we recommend Strawberry Perl 5.32.1.1; on Linux, Perl is typically installed from your Linux distribution’s repository. Mascot Parser provide routines for handling modifications with Unimod and reading MSP files. The Mascot Parser licensing allows a developer to distribute code and applications that use the library as long as they do not charge any license fee for the application. Commercial licenses are also available.
Download the converter and install Mascot Parser as per the included readme instructions. Once this is done, it is simple to convert between BLIP and MSP formats on the command line:
# BLIB to MSP (auto-detected from extension)
perl blib2msp.pl -i library.blib
Or
# MSP to BLIB with optional output file name:
perl blib2msp.pl -i library.msp -o output.blib
Modifications
At the end of a SL conversion, a table of modifications in the library is reported:
When searching one or more SLs, but no FASTA databases, Mascot automatically uses modifications from the spectral library, so you don’t need to specify fixed or variable modifications in the search form.
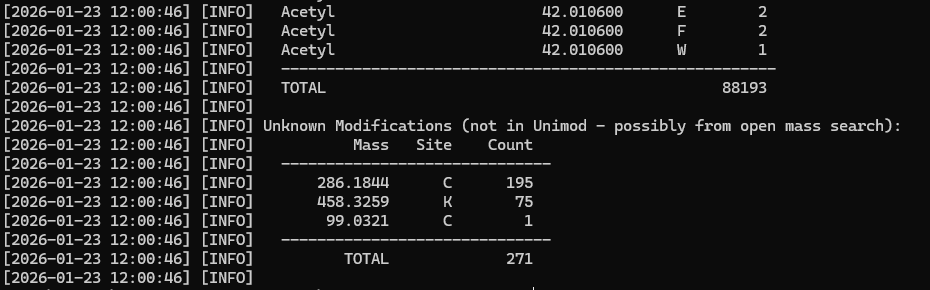
If the BLIB SL was created from an open mass search or uses modifications not included in Unimod, the converter will report these as masses:
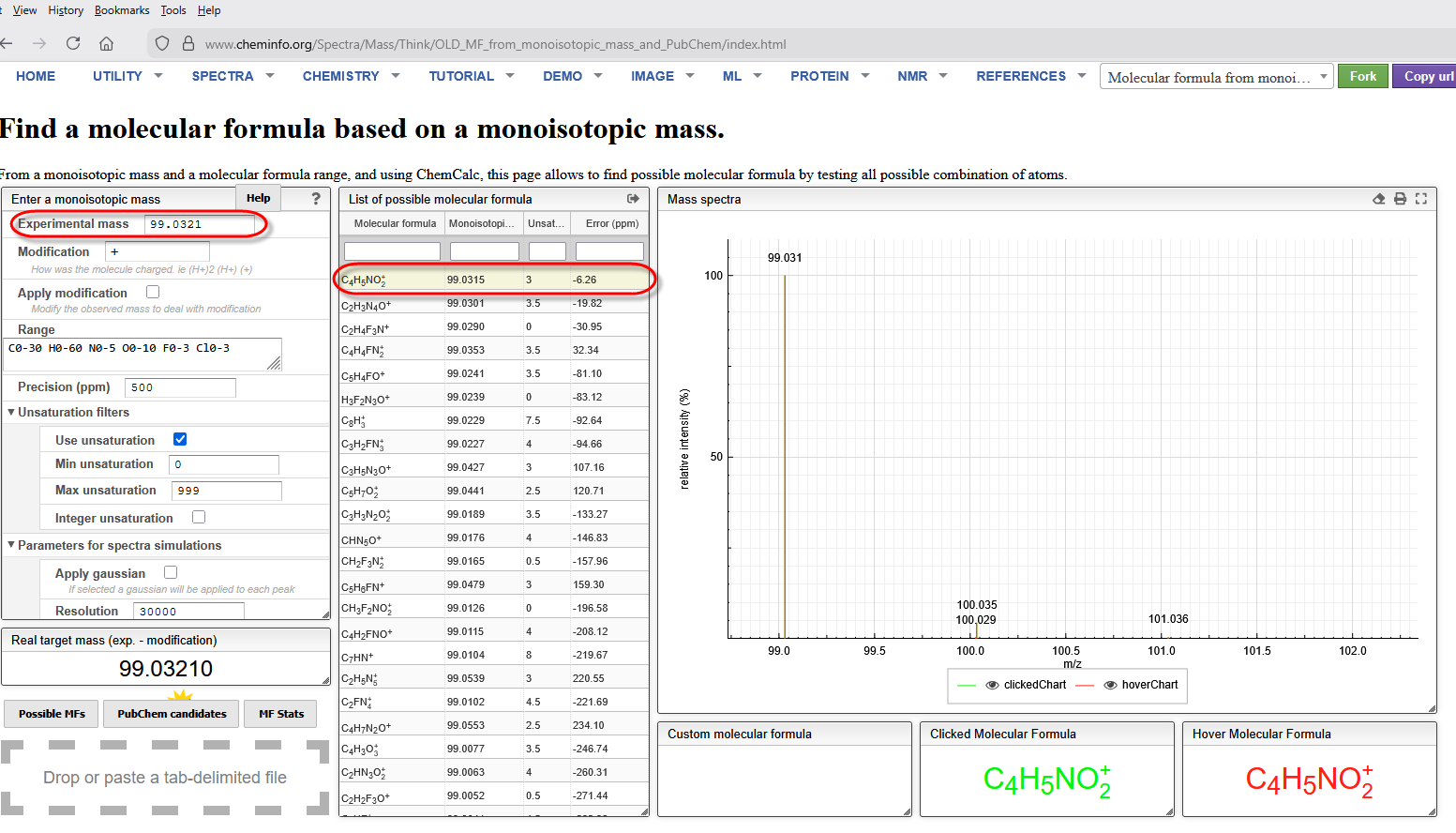
We may not know what these masses are, but we need to add them to Mascot Server to bring the SL on line. The first two we can figure from the masses 286.1844 on Cys is equivalent to TMT6plex + Carbamidomethyl, while 458.3259 on Lys is TMT6plex on the N-term and side chain. You’ll need a likely molecular composition for the mass and add them as a custom modification to our Mascot Server. For the unknown 99.0321 on Cys We used the ChemCalc tool at Cheminfo.org to create a list of candidate chemical compositions and then selected the nearest one by mass error.
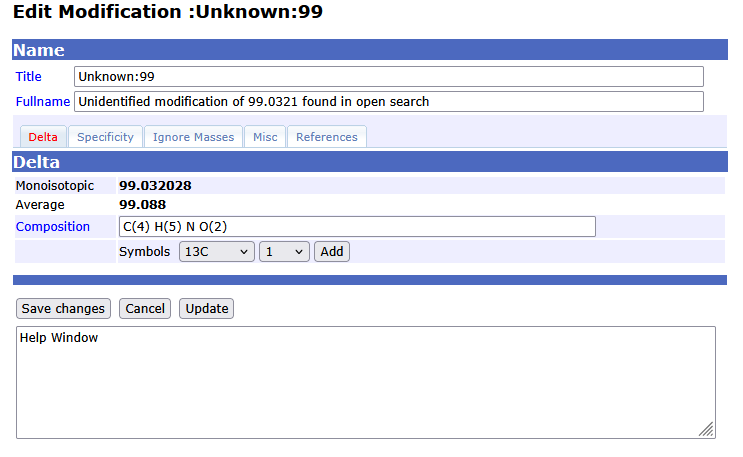
Select the entry then right click and choose Export→selected for pop up window with the formula that can be copied to the clipboard. In a browser, open Mascot Server→Configuration editor→ Modifications editor, create a new modification, then paste the formula into the Composition field. Edit the composition by putting parentheses around the element counts.
Add the modification site amino acid in the specificity tab, then save. I recommend using the same naming format as the other unknown modifications detected by open searches, for example Unknown:99 rather than 99 flake which is what I was thinking of.
As the BLIB library was not converted using the newly updated unimod file, you either need run BLIB to MSP conversion again using a copy of your local Mascot Server mascot\config\unimod.xml file, or add aliases for the masses to mascot\config\library_mod_aliases:
"286.1844" = "TMT6plex + Carbamidomethyl" "458.325" = "Double TMT6plex" "99.0321" = "Unknown:99"
My preferred approach would be to convert the library again although the final results are identical.
Working with mixed species libraries
When you have a SL generated from samples containing multiple organisms, a metaproteomic SL, there are a couple of additional features that can be useful. One example of such a metaproteomic SL would be a library generated from the analysis of a swab of an infected wound that is cultured in media containing BSA and other proteins. Such a sample would contain proteins from the infected organism (human/animal), the infecting bacteria and the culture media.
When you bring a library online in Mascot Server, it will automatically map proteins to the FASTA database specified in the SL definition. You can also use the protein mapping feature --fasta-dir at library creation time. The one advantage of doing this at the SL level is that you can use multiple databases that cover bacteria, human and BSA proteins in the mapping process. You will need to install some additional Perl modules to carry out the mapping (instructions in the associated readme file). Note, however, including species mapping will increase the time taken to run the conversion.
There are also two utility scripts that can be useful for metaproteomic analysis. The first script creates a list of peptide sequences in the database. And the second script will filter a database so that it only contains those spectra. Their usage will be discussed in a follow up article.
Keywords: metaproteomics, spectral library