Identify more proteins using Mascot and Proteome Discoverer with this one small trick
Mascot Server is often used with software packages like Thermo Proteome Discoverer™ (PD). We recently made it possible to enable machine learning on the server side by specifying it in the Mascot instrument definition, which is also the topic of one of our ASMS 2025 posters. An important detail was overlooked in the initial instructions, which is: you should disable MudPIT scoring when machine learning is in use. This unusual step is necessary for the Protein FDR Validator node to work correctly in PD.
Problem: why am I not getting more protein hits?
For benchmarking, and for the ASMS poster, we used PRIDE project PXD028735. This is the raw data for A comprehensive LFQ benchmark dataset on modern day acquisition strategies in proteomics (Pyuvelde et al., Scientific Data, 9(126), 2022).
We selected raw files for conditions A and B for sample alpha, which were run in four technical replicates on Thermo Orbitrap QE HF-X. The paper actually mentions only triplicate measurements, so we used the technical replicates 1, 2 and 3. Conditions A and B differ by the mixture of human, yeast and E. coli.
After processing the files in Proteome Discoverer 3.2 and Mascot Server 3.1, a curious pattern emerged. Enabling spectrum prediction (MS2PIP) and RT prediction (DeepLC) produced more peptide matches, compared to just using the Percolator node, but Proteome Discoverer did not identify any more proteins. Compare the protein counts in the table below.
| No ML | Percolator node | Mascot ML | ||||
|---|---|---|---|---|---|---|
| Instrument | ESI-TRAP | ESI-TRAP | HCD2019:hela_lumos_2h_psms | |||
| #proteins | #peptides | #proteins | #peptides | #proteins | #peptides | |
| Identified | 4268 | 40964 | 6785 | 52935 | 6483 | 57098 |
| Quantified | 4241 | 33103 | 6442 | 40341 | 6298 | 42524 |
The count of proteins is from the Proteins tab in PD, and the count of peptides is Peptide Groups, all at the default 1% FDR. The first case (No ML) is without any machine learning. The second case (Percolator node) makes a standard Mascot search, followed by the Percolator node in PD. Finally, the third case (Mascot ML) causes Mascot to refine the results using MS2Rescore and Percolator, then the refined results are sent to PD.
So, enabling MS2PIP and DeepLC (HCD2019:hela_lumos_2h_psms) produces 8% more peptides over the Percolator node, but it fails to translate into more protein hits. What is going on?
Solution: turn off MudPIT scoring
The solution is not what you might expect. In PD, go to Administration, Processing Settings, Mascot, Mascot Protein Score and turn off MudPIT scoring:
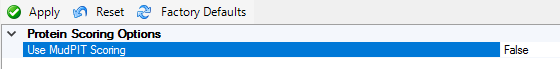
Close and reopen the study, then reprocess the consensus workflow. You don’t need to change any parameters in the workflow itself.
When the processing is done, the counts of peptide groups remain the same, but PD is now able to identify more protein hits.
| No ML | Percolator node | Mascot ML | ||||
|---|---|---|---|---|---|---|
| Instrument | ESI-TRAP | ESI-TRAP | HCD2019:hela_lumos_2h_psms | |||
| #proteins | #peptides | #proteins | #peptides | #proteins | #peptides | |
| Identified | 4268 | 40964 | 6785 | 52935 | 6899 | 57098 |
| Quantified | 4241 | 33103 | 6442 | 40341 | 6566 | 42524 |
Now the 8% more peptide matches translates into 124 more proteins quantified.
Why it works
The explanation is found in the Protein FDR Validator node, which uses either the protein score or a Sum PEP Scores metric for estimating protein FDR.
Percolator node: When the Percolator node processes Mascot results, it calculates a Sum PEP Scores metric. This is -∑ilog10(PEPbest peptide, i), where the best (smallest) PEP is selected for each identified peptide. The Protein FDR Validator node uses Sum PEP Scores when setting a score threshold between target and decoy proteins.
Mascot ML: When Mascot refines the results, it returns -10log10(PEP) as the peptide score. However, the Mascot node does not currently “unpack” the PEP from the peptide score, so the downstream PD nodes are ignorant of the computed PEP. The Protein FDR Validator node falls back to using the protein score when setting a score threshold between target and decoy proteins.
Protein FDR Validator (Standard score): When MudPIT scoring is disabled, Standard protein scoring is used. The standard protein score is just the sum of scores of the peptides assigned to the protein hit, where the best score (smallest PEP) is selected for each identified peptide. If you write it out as a formula, it turns out to be -10∑ilog10(PEPbest peptide, i), exactly equivalent to the Sum PEP Scores. When the Protein FDR Validator node uses the standard protein score, the thresholding is equivalent to Sum PEP Scores. (The factor of 10 is irrelevant.)
Protein FDR Validator (MudPIT score): The MudPIT score was designed as a non-probabilistic ranking of protein hits. It’s a rather complicated formula of PSM scores and score thresholds, and it’s not equivalent to Sum PEP Scores. When the Protein FDR Validator node uses MudPIT scores, it’s unable to produce a good thresholding between target and decoy proteins – even if you give it plenty more peptide matches.
The theory is well and good, but how can you tell the numerical values are OK? In the PD result viewer, go to the Results Statistics tab and scroll down to Sum PEP Scores or Protein Score rows. These are very handy metrics that confirm the theory.
| Minimum | Median | Maximum | Sum | |
|---|---|---|---|---|
| Sum PEP Score | 0.89 | 13.45 | 712.59 | 1,004,382 |
| Standard score (-10log10(PEP)) |
15.01 | 150.69 | 7274.10 | 10,702,554 |
The order of magnitude closely tracks the factor of 10. (The numbers could only be identical if Mascot Server and PD used exactly the same core features with Percolator.)
New recommended PD steps
We have updated the setup instructions accordingly: Using machine learning with Mascot Server 3.1 and Proteome Discoverer (5 pages, 291 kB)
Briefly, step 1 is to configure an instrument in the Mascot config editor, like MS2PIP:HCD2021, which enables refining and choose a model.
Step 2 is to increase the Max MGF Size in the Mascot node configuration, if you haven’t already done so.
Step 3 is to disable MudPIT scoring (as above).
Step 4 is to run the processing workflow as shown below:
![Mascot processing workflow in Proteome Discoverer: [Spectrum Files] to [Spectrum Selector] to [Mascot] to [Target Decoy PSM Validator]](/images/Mascot_PD_processing_workflow_TD_validator.png)
Remember to select the new instrument in the node settings:
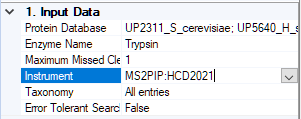
The consensus workflow can be any suitable one of the standard consensus workflows, which all make use of the Protein FDR Validator node.
Keywords: machine learning, MS2PIP, Percolator, Proteome Discoverer