New features in Mascot 2.2
Quantitation
The good news is that Mascot 2.2 contains all the infrastructure needed for a wide variety of quantitation experiments. In some cases, the quantitation results are displayed within a Mascot result report, because all the necessary information is present in the peak list. This applies to the Reporter, (e.g. iTRAQ as shown in the screen shot, below), Multiplex, and emPAI protocols. In fact, emPAI is "always on", and will be reported whenever an MS/MS search contains at least 50 spectra. For a full description of these protocols, and which chemistries they apply to, the starting point is the quantitation help pages.
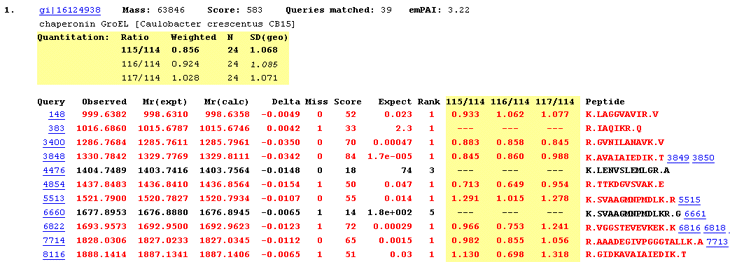
The bad news is that you cannot generate quantitation reports for the other protocols until the Mascot Distiller Quantitation Toolbox is released. This affects the Precursor, Replicate, and Average protocols. Precursor is quantitation based on the relative intensities of extracted ion chromatograms (XICs) for precursors within a single data set. This is by far the most widely used approach, which can be used with any chemistry that creates a precursor mass shift. For example, 18O, AQUA, ICAT, ICPL, Metabolic, SILAC, etc., etc. Replicate is Label free quantitation based on the relative intensities of extracted ion chromatograms (XICs) for precursors in multiple data sets aligned using mass and elution time.
We’re working on getting the Mascot Distiller Quantitation Toolbox released ASAP …
The changes required to support quantitation were quite extensive:
- All the settings for quantitation experiments are encapsulated into named quantitation methods, defined in an XML configuration file, quantitation.xml
- Mascot now takes its modification definitions direct from an XML representation of the Unimod database. In Unimod, both amino acid residues and modifications are defined in terms of their elemental composition.
- Metabolic labelling, in which the isotopic label is present throughout the peptide backbone, is now supported.
- Specialised modifications can be defined locally, within a quantitation method.
- Mascot now supports exclusive modifications. A group of exclusive modifications can be thought of as a choice of fixed modifications. For example, a SILAC experiment might have three or four different labels which will never be mixed on a single peptide.
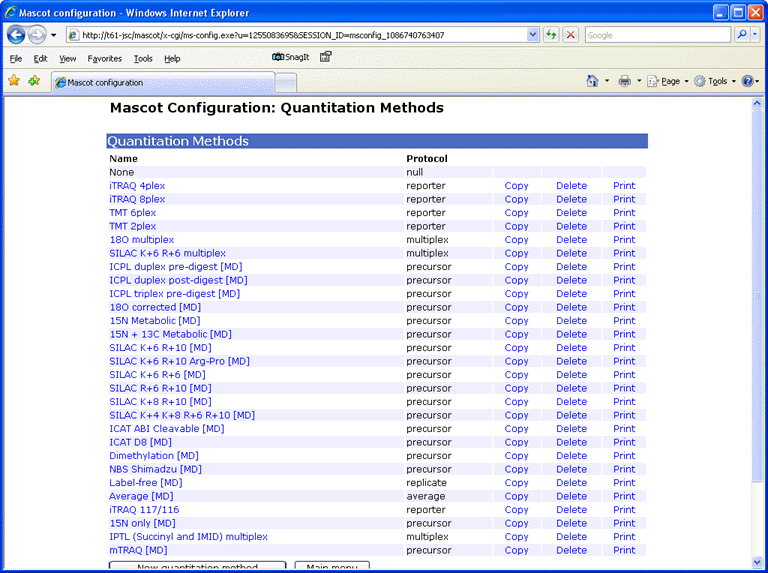
Configuration Editor
A new, web browser based configuration editor can be used to view and edit quantitation.xml, unimod.xml, and the other Mascot configurations files. More details here, and in the Mascot Setup & Installation Manual.
Automatic Error Tolerant Searches
An automatic error tolerant search is performed by choosing the Error Tolerant checkbox on the search form. A standard, first pass search is performed using the search parameters specified in the form. From the results of the first pass search, all of the database entries that contain one or more peptide matches with scores at or above the homology threshold, (or identity threshold if there is no homology threshold), are selected for an error tolerant, second pass search. At the completion of the second pass search, as single report is generated, combining the results from both passes.
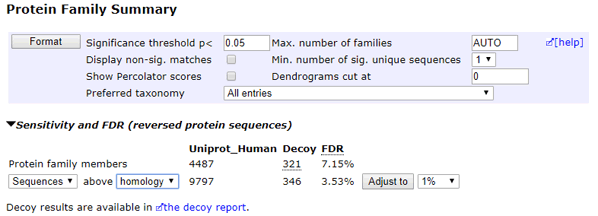
Automatic Decoy Database Searches
For an automatic decoy database search, choose the Decoy checkbox on the search form. During the search, every time a protein or peptide sequence from the "forward" database is tested, a random sequence of the same length is automatically generated and tested. The average amino acid composition of the random sequences is the same as the average composition of the forward database. The matches and scores for the random sequences are recorded separately in the result file. When the search is complete, the statistics for matches to the random sequences, which are effectively sequences from a decoy database, are reported in the result header.
Other important changes
- 64 bit versions of Microsoft Windows are now supported.
- Mass values are now selected, iteratively, by peak intensity for PMF searches. This will give better matches where there are a lot of weak, noise peaks.
- A new search parameter to handle cases where the 13C peak was picked without having to open out the mass tolerance.
- Additional search parameters can now appear in the local scope of a single query, such as instrument type and variable modifications. This is useful for experiments like alternate CID / ETD.
- A new utility, TS2Mascot, for importing peak lists from an Applied Biosystems 4000 series databases.
- Mascot Daemon has a reduced memory footprint and can now handle raw files and peak lists larger than 2 GB.
- Many improvements and bug fixes to the export utility.
- Added z+2H ion series for better ETD matching.
- … and lots more
