Divide and conquer: Fractionated Label Free Quantitation in Mascot Distiller 2.8.2
We have recently released Mascot Distiller 2.8.2. The headline new feature is support for Label-Free Quantitation of fractionated samples.
With Mascot Distiller 2.8, individual raw files in a project are aligned against a consensus generated by roughly aligning and combining the total ion chromatograms (TICs) of each raw file. If a peptide match is found in one sample file and not another, the alignment is then used to calculate a starting point for peptide XIC detection in the file missing the identification. You can find more details about this in a previous blog article.
The issue with fractionated samples is that the generated consensus is an amalgam of all the fractions which will not be an accurate representation as there will be little or no overlap in the peptides between the fractions.
In Mascot Distiller 2.8.2, we have changed this so that for fractionated samples, multiple consensuses are generated – one for each fraction. Files are then aligned to the consensus for their assigned fraction. In addition to enabling proper support for fractionated samples, this makes quantitation faster for these types of experiment as the system doesn’t waste time looking for a peptide in all the files – it only looks for it in the fraction (or fractions) it was identified in.
GelC-MS Example
One of the most common examples of a fractionated LFQ experiment is the ‘GelC-MS’ approach. Here, the protein samples are first run on a 1D Gel, separating the proteins, then the lanes are then cut into a fixed number of equally sized gel slices which are then digested and LC-MS/MS run on the digests. The gel slices are, effectively, separate fractions of the sample.
To demonstrate fractionation support in Distiller 2.8.2, we took the data from a publicly available dataset from the PRIDE repository, PXD029062. This comprises of two samples, from HeLa and HeLa KO30 cells, with protein extracts from each sample run on a separate gel lane. Four equal slices were then taken from each lane and processed, resulting in a total of 8 raw files.
We re-processed and searched the raw files using Mascot Daemon with the Mascot Distiller Daemon Toolbox. Mascot Distiller 2.8.2 was used by Daemon for peak-picking, and the generated peak lists files searched with the “Label-free” quantitation method selected against the UniProt Human reference proteome using Mascot Server 2.8.1
The .rov project files generated by Daemon were then used to create a multifile project in the Mascot Distiller GUI and the data quantified in two different ways:
- Align only matching fractions using the new settings in Distiller 2.8.2
- Align all files against each other, giving the same results as you would get in Distiller 2.8 or 2.8.1
Figure 1 below shows the interface for setting up components and fractions in Distiller 2.8.2. The combination of Component and Fraction must be unique, so we can now have multiple raw files associated with a single component provided they are all given different fraction numbers. If you have a large number of fractions per sample, it could be tedious to enter the fraction numbers manually, so you can instead select a range of cells, then right click to bring up the context menu and click “Add integer series”. This will fill the cells with an integer series from 1 to the number of rows selected.
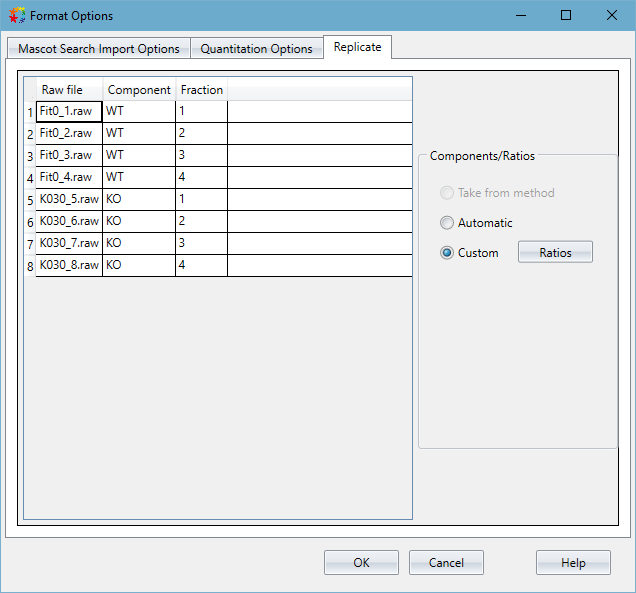
Click to view full size image
Figure 1: The interface for mapping the project raw files to the component and fraction. This is a table with the project raw files in the first column, and the columns for “Component” and “Fraction” which are user editable. Component is the sample ID – for this dataset we have two samples – W.T. and KO – so we’ve entered that into the “Component” column cells for the files as shown. If you have multiple technical or biological replicates of a sample, you should encode that information into the component names.
If you want all the files to be aligned against each other, you should instead leave the “Fraction” column blank. Each component name now needs to be different since the combination of component and fraction must uniquely identify the raw file.
Table 1 shows the overall results of running quantitation on the dataset with either aligning all files together, or just aligning matching fractions. In both cases we’ve quantified a total of 1417 peptides. However, quantitation where we’ve aligned just the fractions took only 11 minutes to complete compared with 26 minutes if we align all files – this is because Distiller doesn’t waste time looking for peptides which were only identified in, say, fraction 3 in the files for fractions 1,2 and 4.
In addition to that, we’re getting better quality results back if we align just the individual fractions with 1137 quantified peptide passing our quality thresholds as opposed to just 569 if we align everything.
| Align Everything | Align Fractions | |
|---|---|---|
| No. peptide ratios | 1417 | 1417 |
| Time taken (min) | 26 | 11 |
| No. valid peptide ratios | 569 | 1137 |
One of the quality thresholds we use in Mascot Distiller is the correlation values between the observed and expected precursor isotope distributions. If we allow Distiller to align everything and search for XICs in all the files, we’re adding a lot of junk and noise to data, reducing the overall correlation. The effect is illustrated in figure 2, where you can see that the number of peptides with a correlation of between 0.75 and 1.0 is increased from ~720 to ~1300 if we align within matching fractions only. Since we have a default correlation threshold of 0.8, aligning just the fractions gives us many more valid ratios.
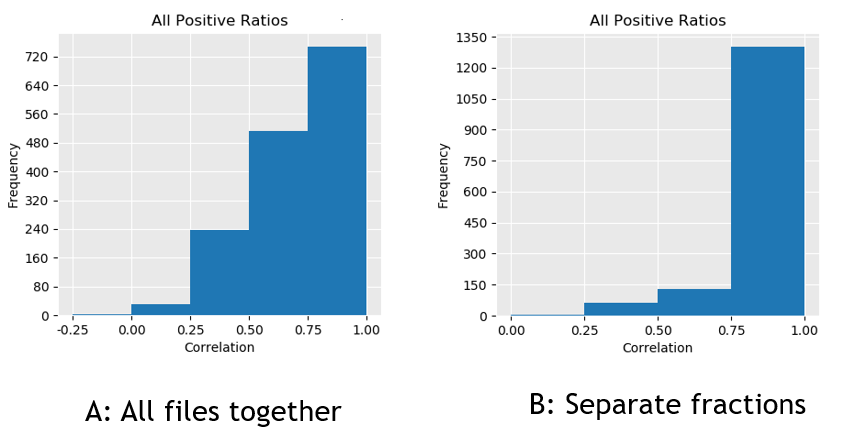
Click to view full size image
Figure 2: Frequency histogram showing the correlation between observed and expected precursor isotope distributions for all quantified peptides where we’ve either (A) aligned all files together or (B) aligned just matching fractions.
If you already have a licence for Mascot Distiller, then 2.8.2 is a free update. If not, and you’d to evaluate it, we offer a 30 day trial of Mascot Distiller. For details, please see the Mascot Distiller download page.
Keywords: label-free, Mascot Distiller, quantitation, replicate